Low mitochondrial DNA diversity of Japanese Polled and Kuchinoshima feral cattle
Abstract
This study aims to estimate the mitochondrial genetic diversity and structure of Japanese Polled and Kuchinoshima feral cattle, which are maintained in small populations. We determined the mitochondrial DMA (mtDNA) displacement loop (D-loop) sequences for both cattle populations and analyzed these in conjunction with previously published data from Northeast Asian cattle populations. Our findings showed that Japanese native cattle have a predominant, Asian-specific mtDNA haplogroup T4 with high frequencies (0.43–0.81). This excluded Kuchinoshima cattle (32 animals), which had only one mtDNA haplotype belonging to the haplogroup T3. Japanese Polled showed relatively lower mtDNA diversity in the average sequence divergence (0.0020) than other Wagyu breeds (0.0036–0.0047). Japanese Polled have been maintained in a limited area of Yamaguchi, and the population size is now less than 200. Therefore, low mtDNA diversity in the Japanese Polled could be explained by the decreasing population size in the last three decades. We found low mtDNA diversity in both Japanese Polled and Kuchinoshima cattle. The genetic information obtained in this study will be useful for maintaining these populations and for understanding the origin of Japanese native cattle.
Introduction
Four breeds of native beef cattle (Wagyu) have been established in Japan: Japanese Black, Japanese Brown, Japanese Shorthorn and Japanese Polled. Japanese Black cattle are the dominant beef breed in Japan and are famous for their high-quality meat. Japanese Brown are categorized into two different substrains: Kochi (Tosa-strain) and Kumamoto (Higo-strain). The Japanese Shorthorn have been improved by crossbreeding with imported Shorthorn bulls to the indigenous native cattle in the northern region of Japan. Japanese Polled have also been improved by crossbreeding with imported Aberdeen Angus bulls and have been maintained in a limited region of Yamaguchi in Japan. These breeds were established by crossing Japanese native cattle with several breeds of European cattle during the mid-19th century to improve the native stock.
In addition to the Wagyu breeds, there are two unique native cattle populations in Japan: Mishima and Kuchinoshima cattle. Mishima cattle have been isolated on Mishima Island for at least 200–300 years and are conserved as a closed colony. Mishima cattle retain the characteristics of native Japanese cattle and were declared a ‘national natural treasure’ in 1928 (Tsuda et al. 2013). Kuchinoshima cattle are unique feral cattle and originated from grazing Japanese native cattle on Kuchinoshima Island in the Tokara Island chain of Kagoshima during the Meiji and Taisho periods (Kawahara-Miki et al. 2011; Siqin et al. 2014). So far, these native cattle have been maintained without any genetic influences from European breeds.
Mitochondrial DNA (mtDNA) is a powerful genetic marker for investigating the origins of livestock. The mtDNA variations in the displacement loop (D-loop) region have been widely used for studying the origin, genetic diversity and relationships in cattle (Loftus et al. 1994; Mannen et al. 1998; Troy et al. 2001; Sasazaki et al. 2006; Chen et al. 2010). Previous studies demonstrated the genetic information and relationships in Japanese Black (Mannen et al. 1998, 2004), Japanese Brown (Sasazaki et al. 2006) and Mishima cattle (Shi et al. 2002) using mtDNA variations.
Japanese Polled and Kuchinoshima cattle are maintained as small populations (< 200) compared with other Japanese breeds and have been subject to significant inbreeding. Knowledge of the basic genetic information, such as mtDNA sequences, is fundamental to the conservation of native cattle populations. The objective of this study is to determine mtDNA variations and genetic diversity in Japanese Polled and Kuchinoshima cattle and to analyze these sequences in conjunction with previously published mtDNA data from Northeast Asian cattle populations to better understand the genetic diversity and structure of Japanese Polled and Kuchinoshima cattle.
Materials and Methods
Ethical conditions
All procedures in the present study were performed according to the Research Guidelines for Kobe University.
Animals
Genomic DNA was extracted from blood samples of 57 Japanese Polled and 32 Kuchinoshima cattle. Kuchinoshima cattle have been maintained in Kagoshima University (13 animals used in this study) and Nagoya University (19 animals). The Kagoshima and Nagoya populations have originated from captured seven males and five females, and four males and six females, respectively (personal record in Kagoshima and Nagoya Universities). Since the relationships were unclear, we used maximum available number of the cattle (32 Kuchinoshima) in this study. Representative published mtDNA sequences and genetic information were included to construct the phylogenetic tree; 32 Japanese Black (Mannen et al. 1998; accession nos. U87633‑U87650), 60 Japanese Brown (30 Kumamoto and 30 Kochi strains) (Sasazaki et al. 2006; AB244486‑AB244514) and two Mishima cattle (Shi et al. 2002; AB177788, AB177789). In addition, the sequences and genetic information of 30 Korean native cattle (Hanwoo) (Mannen et al. 2004; AB117037-AB117059) and 48 Mongolian native cattle (Mannen et al. 2004; AB117060-AB117092) were used to construct reduced median networks.
Sequencing
We amplified the complete D-loop region of mtDNA using primers constructed from cytochrome b (5′-ACAACTAACCTCCCTAAGACTC-3′) and 12S ribosomal RNA (rRNA) (5′-GATTATAGAACAGGCTCCTC-3′) gene sequences. The mtDNA amplification and sequencing were performed according to previous studies (Mannen et al. 2004). Variations in the D-loop region of Japanese Black were defined by comparison with the reference bovine mtDNA sequence (accession no. V00654) published by Anderson et al. (1982).
Sequence analysis
Sequence alignment of the D-loop region was performed using CLUSTAL W (Thompson et al. 1994). To investigate the genetic relationship among mitochondrial sequences, an un-rooted neighbor-joining phylogenetic tree (Saitou & Nei 1987) was constructed using the Tamura-Nei distance (Tamura & Nei 1993). The distance computation and phylogenetic tree construction were incorporated into the MEGA package Ver. 5.03 (Tamura et al. 2011). All sites containing alignment gaps were excluded from the analysis. Reduced median networks were constructed using NETWORK 4.5 (Bandelt et al. 1995).
Results
We analyzed the complete D-loop sequences of 57 Japanese Polled and 32 Kuchinoshima cattle, and deposited these sequences in the DNA Data Bank of Japan (DDBJ) database (accession nos. LC013966‑LC013978). Figure 1 indicates the alignment of the D-loop sequence with the representative sequences of Japanese Black, Japanese Brown, Mishima and Friesian cattle (reference sequence: V00654). Comparison of these sequences revealed 40 variants, including one indel, two transversions and 37 transitions. On the basis of these variants, Japanese Polled had 12 mitochondrial haplotypes and Kuchinoshima cattle had one mitochondrial haplotype.
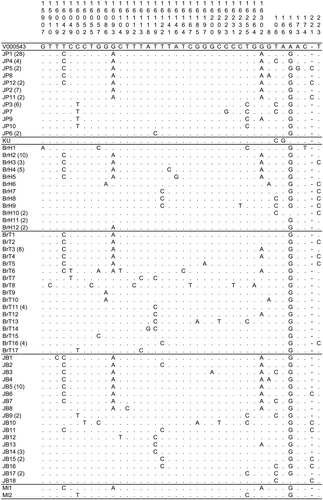
The average sequence divergence values between populations and within populations and the genetic distances among populations are presented in Table 1. Japanese Black, Japanese Brown and Mishima cattle populations displayed similar levels of divergence (0.36–0.55%), while Kuchinoshima and Japanese Polled cattle showed low divergence (0.00–0.20%).
| J. Polled | Kuchinoshima | J. Black | J. Brown-Kumamoto | J. Brown-Kochi | Mishima | |
|---|---|---|---|---|---|---|
| J. Polled | 0.0020 | 0.0056 | 0.0035 | 0.0033 | 0.0041 | 0.0030 |
| Kuchinoshima | 0.0061 | 0.0000 | 0.0058 | 0.0061 | 0.0059 | 0.0060 |
| J. Black | 0.0033 | 0.0057 | 0.0047 | 0.0044 | 0.0047 | 0.0041 |
| J. Brown-Kumamoto | 0.0032 | 0.0062 | 0.0038 | 0.0036 | 0.0042 | 0.0038 |
| J.Brown-Kochi | 0.0040 | 0.0062 | 0.0043 | 0.0042 | 0.0044 | 0.0040 |
| Mishima | 0.0031 | 0.0061 | 0.0037 | 0.0039 | 0.0042 | 0.0055 |
- Above the diagonal and on the diagonal are the average sequence divergences between populations and within populations, respectively. Below the diagonal are genetic distances between populations calculated by the Tamura-Nei distance (Tamura & Nei 1993). We used the published mtDNA sequences and the genetic information obtained from previous studies (Mannen et al. 1998, 2004; Shi et al. 2002; Sasazaki et al. 2006).
Figure 2 shows a phylogenetic reconstruction of Japanese native cattle using mtDNA sequences. All Japanese native cattle belonged to the Bos taurus mtDNA haplogroups T1, T2, T3 and T4 defined by Troy et al. (2001). All Kuchinoshima cattle had only one mtDNA haplotype belonging to haplogroup T3.
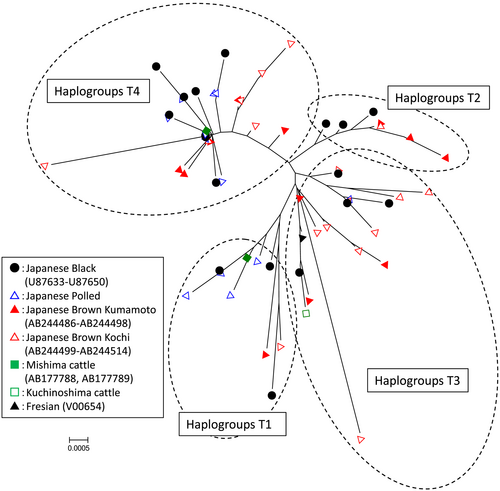
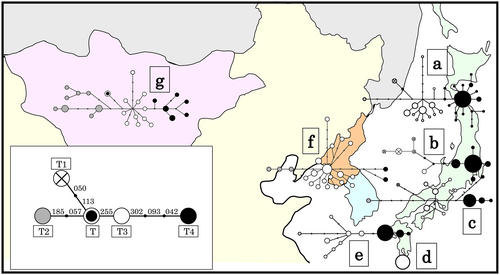
The genetic frequencies of mtDNA haplogroups in Japanese cattle populations are presented in Table 2. Reduced median networks of the mitochondrial haplotypes in the North East Asian cattle are illustrated in Figure 3. All Japanese cattle populations had the Asian-specific mtDNA haplogroup T4 with high frequencies (0.43–0.81), except for Kuchinoshima cattle. The haplogroup T4 consists in a common, phylogenetically central haplotype with derivative sequences that differed by only a few substitutions. Three Japanese Wagyu breeds have a similar mtDNA topology composed by the major haplogroup T4 and the other haplogroups T1‑T3. The Mongolian and Korean networks are composed in several T haplogroups and show high mtDNA diversity (Mannen et al. 2004).
| Population | Mitochondrial haplogroup† | |||
|---|---|---|---|---|
| T1 | T2 | T3 | T4 | |
| J.Polled | 0.16 (9) | - | 0.03 (2) | 0.81 (46) |
| Kuchinoshima | - | - | 1.00 (32) | - |
| J.Black | 0.06 (2) | 0.03 (1) | 0.38 (12) | 0.53 (17) |
| J.Brown-Kumamoto | - | 0.03 (1) | 0.33 (10) | 0.63 (19) |
| J.Brown-Kochi | 0.03 (1) | 0.03 (1) | 0.50 (15) | 0.43 (13) |
| Mishima | 0.50 (1) | - | - | 0.50 (1) |
Discussion
In this study, we have determined the complete mtDNA D-loop sequences of Japanese Polled and Kuchinoshima cattle, and analyzed these sequences in conjunction with previously published data from Northeast Asian cattle populations. With the exception of Kuchinoshima cattle, Japanese native cattle have a predominant, Asian-specific mtDNA haplogroup T4 with high frequencies. By contrast, lower frequencies of the haplogroup T4 were observed in Korean (0.07) and Mongolian (0.12) cattle (Table 2 and Fig. 3).
The haplogroup T4 in Japanese native cattle had relatively low genetic diversity, because one of the haplotypes T4 showed predominance and topological centrality within haplogroup T4 (Fig. 3). These results may reflect the geographical and historical background of Japanese native cattle. The primary ancestral cattle were presumably introduced to the Japanese Islands around the second century A.D. (Mukai et al. 1989). It is believed that a limited number of animals were introduced to the Japanese Islands at that time because of the Japanese sea barrier. This would have caused a pronounced founder effect in maternal mtDNA. The predominance of the haplogroup T4 may originate from the primary mtDNA haplotype in Japanese cattle.
Thirty-two Kuchinoshima cattle had only one mtDNA haplotype belonging to haplogroup T3 (Fig. 2 and Table 2). Kuchinoshima cattle are feral cattle that originate from the grazing Japanese native cattle from Kuchinoshima Island (Siqin et al. 2014). It was reported that a small number of Japanese native cattle were introduced to Kuchinoshima Island in 1918 or 1919 (Hayashida & Nozawa 1964). Some of these cattle then escaped to the mountains and became the founder of the Kuchinoshima feral cattle. So far, this population has proliferated naturally in the mountains of Kuchinoshima Island.
The genetic diversity of Kuchinoshima cattle has been estimated using autosomal polymorphisms. Siqin et al. (2014) reported that four out of six functional gene variants (NCAPG, FASN, SCD, SREBP-1, F11 and MC1R) were monomorphic in 32 Kuchinoshima cattle. Saito et al. (2016) genotyped 54 K single nucleotide polymorphisms in Kuchinoshima, Japanese Black, Japanese Brown and Japanese Holstein cattle, and found that the average minor allele frequency in Kuchinoshima cattle was lower (0.089) than in other populations (0.181–0.251). These results also indicate the extremely low genetic diversity of Kuchinoshima cattle, which may be explained by a founder effect and/or genetic drift in the small population (< 100 animals) that has survived on Kuchinoshima Island for many generations.
Japanese Polled cattle have relatively lower mtDNA diversity (0.0020) than other Wagyu breeds (0.0036–0.0047) (Table 1). Japanese Polled cattle have been maintained in a limited area of Yamaguchi and the population is now less than 200. This breed has experienced serious inbreeding, which has reduced the population size and increased the risk of extinction. The low mtDNA diversity in Japanese Polled cattle may be explained by the decreasing population size in the last three decades.
In conclusion, we have defined the structure and diversity of mtDNA in Japanese Polled and Kuchinoshima feral cattle. Our findings demonstrated low mtDNA diversity in Japanese Polled and Kuchinoshima cattle. Kuchinoshima cattle had only one mtDNA haplotype belonging to haplogroup T3, indicating a strong founder effect. The genetic information obtained in this study will be useful for maintaining these populations and for understanding the origin of Japanese native cattle.
Acknowledgments
We thank Wagyu Registry Association for sampling and data collection. This work was supported in part by JSPS KAKENHI Grant Numbers 23380165 and 16K15025.




