Molecular ontogeny of selected immune-relevant and metabolism-related genes in cod, Gadus morhua during early development
Abstract
This study characterized the expression of some immune- and metabolism-related genes in Atlantic cod, Gadus morhua during early development. Of the 24 genes that were analyzed, five genes were highly expressed in the unfertilized eggs, namely, mcl1 and NR-13, which are apoptosis-related genes; Cu/Zn superoxide dismutase (sod), an antioxidant gene; fortilin (tpt), an antiviral gene and glucose transporter-1 (glut1), a metabolism-related gene. Other genes that had moderate expression levels in the unfertilized eggs include: metallothionein (mt), catalase (cat), heat shock protein70 (hsp70) and glut3. These indicate that there was maternal transfer of transcripts involved in immune defence, e.g. antibacterial and antiviral defence, oxidative stress and apoptosis as well as those involved in metabolism, e.g. glucose transport. At the early larval stage, the number of highly expressed genes in the fish increased to eight. Three genes are involved in apoptosis (bcl2, mcl1 and NR-13); an antibacterial gene, mt; an antiviral gene, tpt and three antioxidant genes, glutathione peroxidase (gpx), hsp70 and sod. Cluster analyses revealed that these genes can be grouped into three categories, namely, those genes that are highly expressed throughout the early developmental phase; those that had moderate expression in most of the time points sampled; and those that had mostly weak or undetected expression during the early life stages. The expression of immune- and metabolism-related genes in cod, even in unfertilized eggs indicates that this fish possesses a well-equipped defence system against pathogens and a well-defined system required for metabolism prior to becoming a fully functional organism.
Introduction
Atlantic cod (Gadus morhua) is a commercially important fish species in the North Atlantic. In recent years, this species has received considerable focus as a candidate for aquaculture, thereby lessening the over-dependence from the wild. Improved techniques for mass production of eggs and larvae are available, but high and sporadic mortalities during the early stages of its development still occur. High mortality in fish during their early life stages can either be due to poor egg quality or infection with various pathogens (Seppola, Johnsen, Mennen, Myrnes & Tveiten 2009). Good egg quality has been equated with low mortality during embryonic development and in the first feeding stage of fish (Bromage & Shepherd 1992). The immunological capacity of the embryos is likely one aspect that can gauge egg and larval quality (Seppola et al. 2009). The immune competence that the fish has acquired during their early life stages could be a significant factor for better survival in later stages or when they are reared in aquaculture facilities. Among the different species of fish, the molecular pathways involved in the immune responses during embryonic development have been well-studied in a model fish species, zebrafish, Danio rerio. Antibacterial responses in zebrafish embryos involve the interferon gamma (IFN-γ) signalling pathway (Sieger, Stein, Neifer, van der Sar & Leptin 2009) and they mount an immune response to lipopolysaccharide (LPS) and bacterial pathogens (Watzke, Schirmer & Scholz 2007; Stockhammer, Zakrzewska, Hegedûs, Spaink & Meijer 2009). The innate immune defences develop first and the adaptive immune system follows later (Stockhammer et al. 2009). The clear distinction in the development and ontogeny of the innate and adaptive immune in zebrafish embryos makes them a suitable model to study fish immune response in the early life stages (Stockhammer et al. 2009).
In fish, maternal factors such as mRNAs, proteins, vitamins and minerals are passed from the mother to the unfertilized eggs and these factors are thought to influence fish egg quality and embryonic health (Bobe & Labbé 2010). These factors are important for the developing embryo until such time when its own transcriptional machinery becomes functional including its immune system. Evidence for maternal transfer of important substances in fish eggs that are important in immune defence includes the transfer of maternal immunoglobulins to eggs in some species of fish (Kanlis, Suzuki, Tauchi, Numata, Shirojo & Takashima 1995; Takemura & Takano 1997; Huttenhuis, Taverne-Thiele, Grou, Bergsma, Saeij, Nakayasu & Rombout 2006). In rainbow trout, Oncorhynchus mykiss, the abundance of the prohibitin 2 transcript was inversely related with normal early development of the fish (Bonnet, Fostier & Bobe 2007). G-lysozyme transcripts were detected in unfertilized eggs of rohu, Labeo rohita, whereas transferrin and TLR22-like transcripts were expressed from 6 h post fertilization (Nayak, Mohanty, Mishra, Rauta, Das, Eknath & Sahoo 2011). In Atlantic cod, evidence of maternal transfer of immune-related transcripts such as lysozyme and cathelicidin was observed in unfertilized eggs (Seppola et al. 2009). Furthermore, lysozyme activity was detected in unfertilized eggs, indicating the presence of a functional protein. Analyses of additional gene categories during the early development of cod also revealed the expression of genes related to immunity, oxidative stress, apoptosis and stress (Lanes, Fernandes, Kiron & Babiak 2012). They found that cod eggs obtained from the wild had more abundant transcripts of glutathione peroxidase (gpx) than the eggs obtained from the hatchery. On the other hand, eggs obtained from spawners reared in the hatchery had higher expression of hsp70 than those obtained from the wild. Rise, Hall, Alcock and Hori (2012) also detected several virus responsive and antimicrobial peptide-encoding transcripts in unfertilized cod eggs and at the 2-cell stage. These include, Adam22, hamp, il8, irf1, irf7, lgp2, sacsin and stat1.
At the cellular and protein levels, the ontogeny of the innate immune system in Atlantic cod during the early life stages has been studied by Magnadóttir, Lange, Steinarsson and Gudmundsdóttir (2004). Maternal IgM and C-reactive protein were not detected in fertilized eggs but the complement component C3 and the apolipoprotein A-I were present during organogenesis. Majority of the protein samples in the eggs were vitellogenin-derived maternal proteins. Cathepsin was present during embryonic development but other enzyme activities were restricted to larval samples, especially during first feeding. Furthermore, they found that haemoglobin was detected only at 10 days after hatching.
The development of immune response in Atlantic cod at the molecular level has been given significant attention in previous studies especially in relating it with good quality larvae. However, there are other aspects in embryonic development that need to be studied and the molecular mechanisms that govern those responses have to be elucidated. Hence, in this study we determined the expression profiles of genes related to metabolism particularly in glucose transport during the early development of Atlantic cod. This study also determined the expression profile of other immune-related genes in the fish that were not included in previous studies.
Materials and methods
Sampling of eggs and larvae
Cod eggs and larvae were obtained from a previous study (Ruangsri, Salger, Caipang, Kiron & Fernandes 2012). Unfertilized eggs were collected as well as embryos at different developmental stages (1-cell, 2-cells, 16-cells, oblong, germ ring, 50% epiboly, 10-somite and golden eye) and larvae (hindgut stage, first feeding and 20 days post hatch) were used for the expression analyses. Pooled egg samples (ca. 10 eggs) and individual larvae were kept at −20°C until extraction of total RNA.
Isolation of total RNA and RT-PCR
Total RNA was extracted from eggs, embryos and larvae following standard procedures. First strand complementary DNA (cDNA) was synthesized from the total RNA (1 μg) using qScript™ cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD, USA). The cDNAs were quantified and normalized at a concentration of 10 μg mL−1 with 1 × TE buffer. The samples were stored at −20°C until use for subsequent PCR reactions. The expression of selected genes that are related to six categories: antibacterial activity (bactericidal/permeability-increasing protein/lipopolysaccharide-binding protein, BPI-LBP; hepcidin, hamp; metallothionein, mt; and, transferrin, tf), antiviral response (fortilin, tpt; ISG15; IRF-1, irf1; and methyltransferase, mett), cytokine production (IL-8, il8; IL-10, il10; IL-1β, il1b; and IFNγ, ifng), apoptotic activity (Bcl-1; Bcl-2, bcl2; Mcl-1, mcl1; and NR-13), oxidative stress response (catalase, cat; glutathione peroxidase, gpx; Hsp70, hsp70; and Cu,Zn-SOD, sod) and glucose transport (Glut-1, glut1; Glut-2, glut2; Glut-3, glut3; and Glut-4, glut4) was determined using the published primers in previous studies (Caipang, Brinchmann, Berg, Iversen, Eliassen & Kiron 2008; Caipang, Brinchmann & Kiron 2008; Caipang, Brinchmann & Kiron 2009; Caipang, Lazado, Brinchmann & Kiron 2010; Caipang, Lazado, Brinchmann, Rombout & Kiron 2011; Caipang, Hynes, Puangkaew, Brinchmann & Kiron 2008).
PCR amplification was carried out at the following conditions: an initial denaturation at 95°C for 3 min, followed by 32 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s and elongation at 72°C for 1 min and a final elongation step at 72°C for 5 min to complete the reaction. The PCR conditions for the amplification of all genes have been tested at different amplification cycles ranging 28–37, and the optimum value using 32 cycles was used for this study. At this stage, the expression of all genes was at the exponential stage as shown by the varying intensities in the gel electrophoresis. Five microlitres (5 μL) were used for electrophoresis on a 1% agarose gel stained with SYBR Safe™ (Molecular Probes, Carlsbad, CA, USA). The intensity of the band on the image was visualized using a gel documentation system (Eastman Kodak, New Haven, CT, USA). Gene expression was measured using Image J (http://rsb.info.nih.gov/nih-image/) and the expression of each gene transcript relative to the expression of the reference gene, β-actin was calculated. The intensity values based on the Image J analyses that were derived for β-actin in each stage during early development were not statistically different, hence, it was considered as the reference gene. Four biological replicates of each stage of development were used for the analyses.
Clustal and TreeView analysis
The expression levels of the genes were grouped according to their expression patterns during early development using Cluster Program 3.0 (Clustering library version 1.27) (De Hoon, Imoto, Nolan & Miyano 2004). The results of the cluster analysis were visualized with TreeView program (Version 1.60) (Saldanha 2004).
Data analysis
The data are represented as mean ± SD. The expression levels of the genes were arbitrarily categorized either as low or undetected (expression level < 0.5), moderate (between 0.5 and 1.0) and high (>1.0). Student's t-test for independent samples was used to compare the relative expression level of a particular gene at the unfertilized egg stage and at the early larval phase (20 days post hatch). All significance levels were set at P < 0.05.
Results
Expression of antibacterial genes
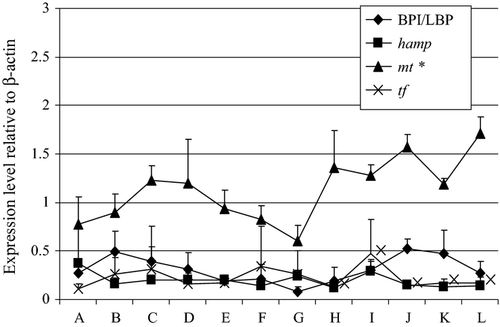
Figure 1 shows the expression profile of selected antibacterial genes in cod during early development. In unfertilized eggs, the genes that were analyzed had low expression levels, except for mt, which had moderate expression levels. The three genes, BPI/LBP, hamp and tr continued to have low expression levels until the first feeding stage of the fish. In contrast to mt that had significantly higher expression level in the 20th day post-hatch larvae than the unfertilized eggs.
Expression of antiviral genes
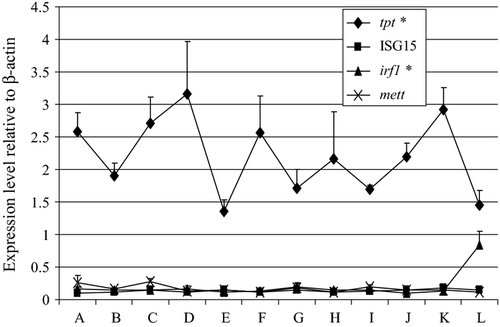
The expression patterns of selected antiviral genes in cod during its early development are shown in Fig. 2. Among the antiviral genes that were analyzed, tpt had high expression levels in unfertilized eggs. This was in contrast to the low or undetected expression levels of ISG15, irf1 and mett. At the 20th day post hatch, irf1 had a significant increase in its expression level. The expression level of tpt was still high during the 20th day post hatch although significantly lower than its expression level in the unfertilized eggs.
Expression of cytokine genes
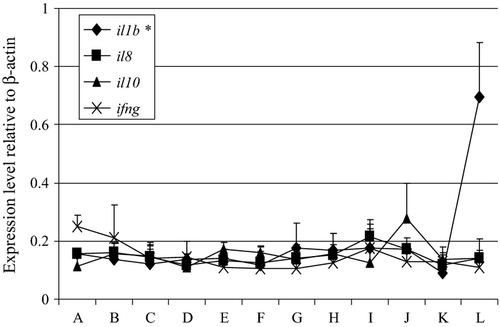
The cytokine genes, namely il1b, il8, il10 and ifng had either undetected or low expression levels in the unfertilized eggs (Fig. 3). Except for il1b, which had a moderate increase in its expression level at the 20th day post hatch, all the other cytokine genes had low expression levels during this phase of cod development.
Expression of apoptosis-related genes
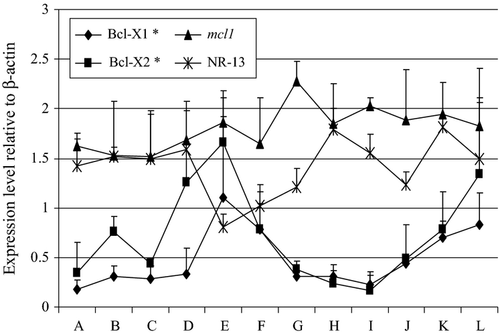
The expression patterns of the different apoptosis-related genes that were analyzed in this study are shown in Fig. 4. There was high expression of mcl1 and NR-13 in unfertilized eggs and the level of their expression continued until the 20th day post hatch. The expression of bcl1 and Bcl-2 significantly increased during the 20th day post hatch in comparison with their levels at the unfertilized egg stage. Bcl-1 and bcl2 were moderately and highly expressed at 20th day post hatch respectively.
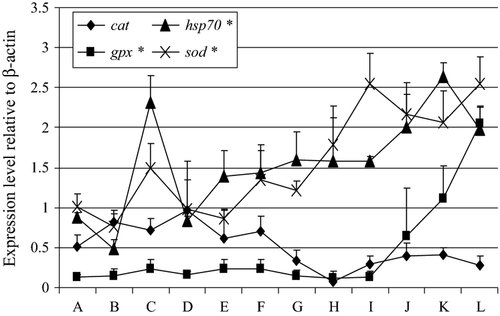
Expression of oxidative stress-related genes
Figure 5 shows the expression patterns of the selected stress-related genes in the embryonic and post-embryonic development of cod. Both cat and hsp70 were moderately expressed in unfertilized eggs, whereas there was high expression of sod. At the 20th day post hatch, gpx, hsp70 and sod were highly expressed at that stage and their levels were significantly higher than their corresponding expression during the unfertilized egg stage. The level of cat expression did not vary during the embryonic and post-embryonic stages of cod.
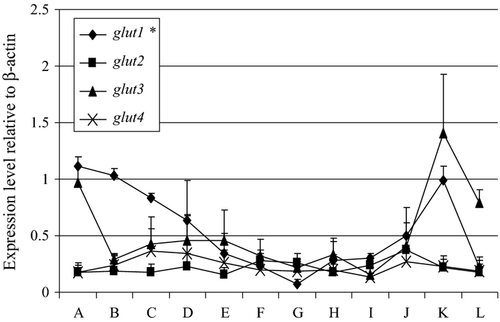
Expression of metabolism-related genes
Figure 6 shows the expression levels of the different metabolism-related genes in the early developmental stage of cod. There was high expression of glut1 in the unfertilized eggs, but its level of expression significantly decreased to become weakly expressed at the 20th day post-hatch stage. Glut3 was moderately expressed at both the unfertilized egg and 20th day post-hatch stages of cod. Both glut2 and glut4 had low expression levels at the unfertilized egg stage and continued until the 20th day post hatch.
Cluster analysis
Using Cluster analysis, the expression patterns of the different genes that were used in the study could be grouped into three clusters (Fig. 7). Cluster 1 includes the genes that had high expression levels from the unfertilized eggs until the 20th day post-hatch stage of cod. The genes in this cluster are composed of mcl1, NR-13, tpt, mt, hsp70 and sod. Cluster 2 is composed of genes that had moderate expression levels during the early developmental stages of cod. The genes that are in this cluster are the following: Bcl-1, bcl2, glut3, cat, BPI/LBP and glut1. The genes that were undetected or had generally low expression levels during the early development of cod are included in Cluster 3. These genes are gpx, tr, glut2, glut4, hamp, mett, il8, ifng, ISG15, il10, irf1 and il1b.
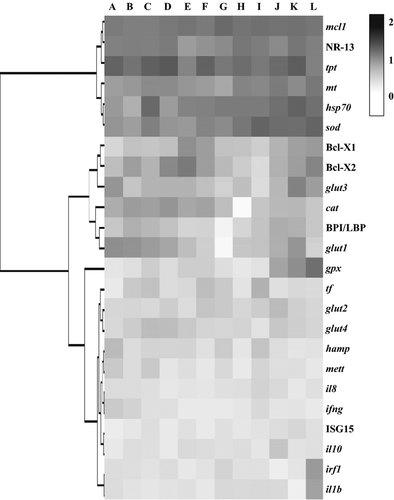
In terms of the stage in cod development, mcl1, NR-13, sod, tpt and glut1 were highly expressed in unfertilized eggs. While bcl2, mcl1, NR-13, mt, gpx, hsp70, sod and tpt had high expression levels during the 20th day post-hatch stage.
Discussion
In this study, we determined the expression profiles of some immune- and metabolism-related genes in the embryonic and post-embryonic stages of Atlantic cod. The expression level of each gene in each stage of development can be considered as the basal level and used as a reference value for subsequent experiments in the early stages of this fish that will be subjected to various treatments. Prior to fertilization, the expression of genes in the eggs of Atlantic cod represents maternal transcripts that were directly transferred from the mother to the eggs. Several studies in cod have indicated maternal transfer of various transcripts related to immunity and metabolism in the fish. In a previous study by Seppola et al. (2009), they found the expression of cathelicidin, cathl and lysozyme, lyzg in unfertilized eggs, indicating maternal transfer of transcripts related to bacterial defence. Lanes, Fernandes et al. (2012) were able to detect eleven genes that are related to apoptosis, immune response and oxidative stress in cod during the fertilization stage. Rise et al. (2012) observed low expression levels of several transcripts that are related to both antibacterial and antiviral responses in unfertilized eggs and at 7 h post fertilization. These include Adam22, hamp, il8, irf1, irf7, lgp2, sacsin and stat1. In this study, we have also observed maternal transfer of transcripts related to apoptosis, oxidative stress, antiviral response and metabolism. It is believed that maternal transfer of immune-related transcripts or proteins to the unfertilized eggs is necessary because these could serve as a first line of defence against various pathogens. However, the expression of other transcripts that have roles in development and metabolism, provides valuable insights that unfertilized eggs as well as the early stages of embryonic development need these transcripts and their protein products for proper development.
Innate immune response in cod is presumed to be active during the early stages of cod development (Seppola et al. 2009). Several transcripts related to antibacterial and antiviral response are expressed in unfertilized eggs and subsequently assume high levels of expression during the early larval stage (Seppola et al. 2009; Lanes, Fernandes et al. 2012; Rise et al. 2012). In this study, we detected high levels of expression of an antiviral gene, fortilin (tpt) in unfertilized eggs, while both tpt and metallothionein, mt were highly expressed during the early larval stage. Fortilin has been found to be involved in the antiviral response in aquatic organisms particularly in shrimp (Graidist, Fujise, Wanna, Sritunyalucksana & Phongdara 2006; Tonganunt, Nupan, Saengsakda, Suklor, Wanna, Senapin, Chotigeat & Phongdara 2008). In humans, tpt has an anti-apoptotic function and is involved in various cellular activities, e.g. growth, division and transformation (Bommer & Thiele 2004). The high expression of this gene in unfertilized eggs of cod likely indicates a dual function for this gene during this stage. It could protect the cells from undergoing programmed cell death, which takes place later in development and at the same time it could have an antiviral role considering that unfertilized eggs are highly susceptible to viral infections in the wild. At hatching and during the early larval stage, both tpt and mt have high expression levels. Fortilin will likely have an antiviral role as well to aid in the maintenance of stable cellular processes. On the other hand, metallothionein is likely to have an antibacterial role. This gene is important in the immune system of the fish because it maintains the homeostasis of essential metals during bacterial infections (Viarengo & Nott 1993) as well as protects the host from cellular damage caused by oxidative stress (Haq, Mahoney & Koropatnick 2003). In mammals, mt aids in sequestration of free metals, thus, depriving the bacteria of the necessary components required for their growth. In fish, mt is also believed to have that function because it has been used as a marker of Flavobacterium psychrophilum infection in rainbow trout, Oncorhynchus mykiss (Orieux, Douet, Hénaff & Bourdineaud 2013). In the skin of naive adult cod, there was high expression level of mt, suggesting its role against bacterial infections (Caipang et al. 2011). Also exposure of the gill epithelial cells of the adult cod with Vibrio anguillarum and Aeromonas salmonicida triggered the expression of mt. Hence, it is not surprising that the early larval stage of cod would rely on mt for its defence against bacterial infections either by regulating the availability of essential metals that are required for bacterial growth or through the production of oxygen radicals that have antibacterial properties. The high expression of both genes in cod during the early stages of development is probably maintained until the fish reaches the pre-adult/adult stage as shown by the high expression levels of both genes in the skin of naive adult cod (Caipang et al. 2011).
Antioxidative enzymes including Cu/Zn superoxide dismutase, catalase and glutathione peroxidise protect the cells from oxidative damage due to the production of reactive oxygen species. The genes responsible for the production of these enzymes are differentially regulated during the early development in rainbow trout at medaka, Oryzias latipes (Fontagné, Lataillade, Brèque & Kaushik 2008; Wu, Shariat-Madar, Haron, Wu, Khan & Dasmahapatra 2011). Lanes, Fernandes et al. (2012) also observed differential regulation of these antioxidative genes during the early stages of cod development. In unfertilized eggs, we have seen high expression of sod and moderate expression level of cat. Our results were similar to Lanes, Fernandes et al. (2012) where they observed high expression levels of these genes at the fertilization stage. A number of genes related to oxidative stress were expressed in cod eggs. This is likely because cod eggs contain a significant amount of polyunsaturated fatty acids (Lanes, Bizuayehu, Bolla, Martins, Fernandes, Bianchini, Kiron & Babiak 2012) and the higher production of the antioxidant is crucial in protecting the embryos against peroxidation (Lanes, Fernandes et al. 2012). Most of these genes producing the antioxidative enzymes are still highly expressed when the cod are in the early larval stages suggesting that these enzymes are needed for efficient metabolism in the developing larvae especially when they shift from endogenous to exogenous feeding, where higher oxygen demands are needed (Rudneva 1999).
The expression of hsp70 was moderate in unfertilized eggs but highly expressed at the early larval stage. Heat shock proteins act as molecular chaperones during embryonic development because they are involved in cell movement, proliferation, morphogenesis and apoptosis (Rupik, Jasik, Bembenek & Widłak 2011). The expression pattern of hsp70 that we observed in this study corroborates the results obtained by Lanes, Fernandes et al. (2012). The fluctuations in the expression of hsp70 in cod during early development as observed in this study could likely indicate the importance of that gene at a particular stage where some cellular processes are mostly affected by hsp70.
During early development in multicellular organisms, the damaged, unwanted and misplaced cells are continuously replenished through apoptosis (Krumschnabel & Podrabsky 2009). However, this process does not take place at any time during embryonic development such that in zebrafish, programmed cell death does not take place before it reaches the gastrula stage (Hensey & Gautier 1997; Ikegami, Hunter & Yager 1999). There are certain gene families that regulate apoptosis and an example of such group is the Bcl-2 family of genes (Borner 2003). This family of apoptosis regulators possesses both pro- and anti-apoptotic genes. In this study, we examined the expression of the anti-apoptotic genes, Bcl-1, bcl2, mcl1 and NR-13. These genes have been previously identified in cod (Feng & Rise 2010) and they bind and sequester the pro-apoptotic Bcl-2 proteins, thus preventing mitochondrial membrane permeabilization (MMP)- induced apoptosis (Brunelle & Letai 2009). We observed high levels of mcl1 in the unfertilized eggs and this was maintained until they reach the early larval stages. Our findings were consistent with the earlier results of Lanes, Fernandes et al. (2012). In contrast with zebrafish, the levels of mcl1a were abundant from fertilization until the blastula stage then dropped to its lowest level during gastrulation, indicating the start of embryonic apoptosis (Chen, Gong, Cheng, Wang, Hong & Wu 2000). In addition, we found high expression of NR-13 in the unfertilized eggs, and it is likely that both NR-13 and mcl1 are responsible in preventing MMP-induced apoptosis in cod eggs. By early larval stage, mcl1, bcl2 and NR-13 had high expression levels, whereas Bcl-1 was moderately expressed. The expression pattern of these anti-apoptotic genes suggests that these members of the Bcl-2 gene family work in a coordinated fashion to ensure cellular equilibrium in the developing larvae.
Passive glucose transporters (GLUTs) facilitate the passage of monosaccharides across the plasma membrane of animal cells, a key stage of carbohydrate metabolism (Baldwin 1993). In mammals, there are at least five distinct sugar transporters (GLUT1–GLUT5). In Atlantic cod, there are four putative glucose transporters (GLUT 1-GLUT4) based on bioinformatic analysis and their expression levels have been determined when the fish have been exposed to various external stimuli (Caipang, Brinchmann, Berg et al. 2008; Caipang, Brinchmann & Kiron 2008; Caipang et al. 2011). There was high expression of glut1 in unfertilized eggs while moderate expression of glut3 was observed. Although the utilization of carbohydrates in fish is much lower than that was observed in higher vertebrates (Teerijoki, Krasnov, Pitkänen & Mölsa 2000), some functional assays of GLUTs in salmon demonstrated that there was glucose transport across membranes and in embryos (Teerijoki, Krasnov, Gorodilov, Krishna & Mölsa 2001). In rainbow trout, glut1 was expressed at the blastula stage until the somite stage (Teerijoki et al. 2001), and the expression of glut1 in unfertilized eggs of cod is consistent with what was observed in rainbow trout. In addition, we have observed that the expression of glut1 in cod decreased as development progresses, which was also similar to the findings obtained by Teerijoki et al. (2001) in rainbow trout. It is likely that glut1 has an important role in glucose transport during the early stages of development and slowly loses its function as the fish grows. This needs further studies and functional assays to determine the exact role of GLUTs in the glucose metabolism in cod.
In conclusion, this study has shown several genes related to immunity and metabolism was expressed in unfertilized eggs of Atlantic cod, indicating maternal transfer of these transcripts. These transcripts have varied roles either by protecting the developing embryos against pathogens or helping in the maintenance of an efficient cellular process in the course development. Regardless of their putative functions, these genes can be grouped into categories based on their expression levels in cod development. Their expression at each stage during the early development in cod might correlate to their degree of importance. Thus, the genes that were analyzed in this study were differentially regulated during the early stages of cod development. These differences are crucial in maintaining a well-coordinated physiological status in the developing fish until they become a fully functional organism.
Acknowledgements
Part of this study was supported by the research funds of the Faculty of Biosciences and Aquaculture, University of Nordland, Norway. The technical assistance of Carlo C. Lazado is gratefully acknowledged. The authors confirm no conflict of interest in this article.




