Multiplex PCR for respiratory bacteria in acute care
Abstract
The purpose of the study was to evaluate the clinical utility of multiplex PCR for detecting bacterial respiratory pathogens in nasopharyngeal samples. Acutely ill adults in the emergency department with respiratory infection symptoms, fever, chest pain or poor general condition were enrolled for this cohort study. Samples were stored at –70 °C until being analysed with multiplex PCR for seven respiratory bacteria. Of the 912 patients enrolled, those with positive bacterial samples (n = 130, 14%) were significantly younger than those with a negative finding (55.5 years vs 62.2 years, p < 0.001), and their mean C-reactive protein (CRP) concentration was higher (110 mg/L vs 59 mg/L, p < 0.0001). Patients with a positive respiratory bacterial finding had a higher probability of pneumonia (35% vs 13%, p < 0.001) and a higher likelihood of receiving a prescription for antibiotics than those with a negative finding (79% vs 59%, p < 0.0001). Positive detection of Streptococcus pneumoniae was associated with a 4.5-fold risk of pneumonia in a multivariate model and detection of an atypical respiratory pathogen with a 9-fold risk. Bacterial PCR performed on nasopharyngeal samples appeared to offer a valuable addition to the diagnostics of infections in adults in acute care.
Respiratory infections are a major cause of hospitalization, antibiotic use and deaths [1, 2]; however, it is difficult to differentiate between viral and bacterial pathogens through symptoms and signs alone in clinical practice [3]. To address this issue, fast and accurate multiplex PCR-based diagnostic tools have been developed for detecting the most probable pathogens.
However, the clinical value of multiplex PCR testing for viruses in acutely ill adults remains unclear. PCR testing using FilmArray RP has been reported to reduce the length of stay in hospital, the duration of antimicrobial administration and the number of chest radiographs performed among patients diagnosed with influenza but not with other viruses [4]. In an open-label, randomized ResPOC study, the length of stay was reduced for a mean of 1.1 days in the active point-of-care viral diagnostics group compared to the routine clinical care group, but there were no differences in antimicrobial use or isolation days [5]. Elsewhere, the rapid testing of multiple respiratory pathogens was found to improve antimicrobial stewardship in patients with pneumonia [6, 7]. On the other hand, in our previous randomized controlled study, the use of multiplex PCR for respiratory viruses did not reduce antimicrobial consumption in acutely ill adults [8]. There are limited data on the values of the commercially available multiplex PCR platforms for assessing respiratory bacterial pathogens in nasopharyngeal swab samples [9], and in any case, many previously evaluated respiratory multiplex PCR panels have lacked a means of detecting Streptococcus pneumoniae and Haemophilus influenzae, which are important aetiological agents in respiratory tract infections.
We had stored nasopharyngeal respiratory samples from our earlier randomized investigation into the use of multiplex PCR for viral pathogens [8], to test the method with bacterial pathogens in this cohort study. Here, we report on the clinical utility of multiplex PCR for detecting seven bacterial respiratory pathogens, including S. pneumoniae and H. influenzae, in acutely ill adults.
METHODS
Study design
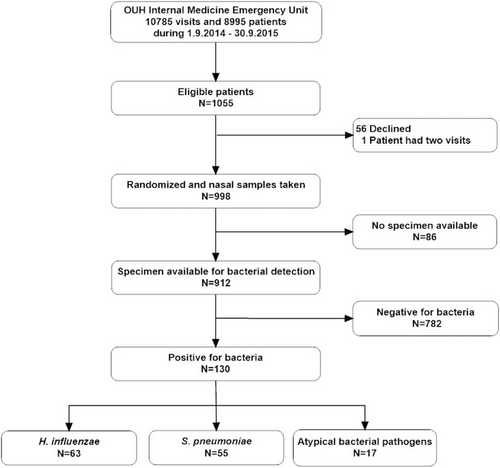
This analysis of the clinical utility of a multiplex PCR assay for respiratory bacteria was based on our previous randomized controlled trial regarding viral diagnostics [8]. In short, we enrolled patients older than 16 years visiting the Internal Medicine Emergency Clinic at Oulu University Hospital who had either (a) respiratory infection symptom such as cough, rhinitis, shortness of breath or sore throat; (b) fever (>38 °C); (c) chest pain or (d) poor general condition for some unknown reason (Fig. 1). The study nurse took nasal swabs from eligible patients who had given their informed consent, and the resulting respiratory samples were frozen and stored at −70 °C. During 2018–2019, the samples were thawed and analysed with the Allplex™ Respiratory Panel 4 (Seegene, Seoul, Korea) for seven bacterial respiratory pathogens: H. influenzae, S. pneumoniae, Chlamydia pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, Bordetella pertusssis and Bordetella parapertussis. The time interval indicated that the bacterial multiplex PCR results had not been available to the clinicians at the time of the patients’ hospitalization.
Clinical data collection
The study nurse also collected data on comorbidities, smoking, current medication and current symptoms at the time of enrolment through a questionnaire, in addition to which the patients’ medical records were reviewed after a follow-up time of 30 days. The study physician then collected data on the course of the disease, the length of hospitalization and the duration of intensive care when required. The results of all microbiological tests and radiological examinations were recorded, and the antimicrobial medication was reviewed from the national electronic database, as well as the hospital records, and supplemented with the information provided by the participants. The national population register (Statistics Finland) was used to confirm the number of deceased patients during the 30 days of follow-up.
Statistical analysis
The sample size for the present study was based on the earlier randomized controlled trial [8]. We used the standardized normal deviate (SND) test to compare the proportions of the various symptoms reported by the patients between those with no bacterial pathogen and those with various positive respiratory bacterial pathogen findings and reported the differences in these proportions with 95% confidence interval (95% CI). Due to a low number of positive findings, we combined C. pneumoniae and M. pneumoniae to form one group of atypical bacterial pathogens for the purposes of the analysis. The hospital admission figures refer to the numbers of patients admitted to Oulu University Hospital. Means and standard deviations (SDs) are reported throughout for continuous variables and 95% CI of the differences for comparisons. The mean ages of the participants with negative vs positive bacterial findings were compared using Student's t-test, and multivariate logistic regression adjusted for age and gender was used to evaluate the risk factors for pneumonia and the positive detection of S. pneumoniae. The analyses were performed using IBM SPSS Statistics for Windows version 27 (IBM Corp., Armonk, NY, USA). The figures were drawn using OriginPro 2020 software (OriginLab Corporation, Northampton, MA, USA).
RESULTS
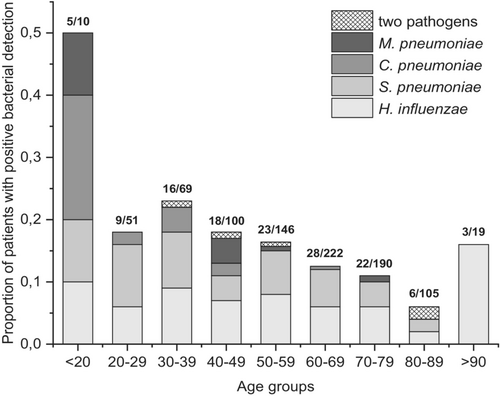
Samples representing 912 patients were available for multiplex PCR assays for respiratory bacteria, and 130 (14.3%) of these patients tested positive for a bacterial pathogen (Fig. 1). The most commonly detected nasopharyngeal pathogens were H. influenzae (63/912, 6.9%) and S. pneumoniae (55/912, 6.0%), while C. pneumoniae was found in 9/912 (1.0%) samples and M. pneumoniae in 8/912 (0.9%) samples. None of the samples tested positive for L. pneumophila, B. pertussis or B. parapertussis. Detection of a bacterial pathogen was most common in patients under 20 years of age, among whom five out of 10 had a positive finding (Fig. 2). The lowest proportion of positive findings was seen in the age group 80–89 years, with 6% (6/105) of the samples positive (Fig. 2).
A respiratory symptom at entry (cough, rhinitis, sore throat or shortness of breath) was reported by 92% (119/130) of the patients with a positive detection result and 82% (641/782) of those without any bacterial pathogen, a difference in 10%, 95% CI 3% to 14%, p = 0.004 (Table 1). As the mean age of the participants was 61.2 years (SD 17.5), those with a positive bacterial detection were significantly younger, with a mean age of 55.5 years (SD 18.4), than those with a negative finding, with a mean age of 62.2 years (SD 17.1), difference of 8.6 years, 95% CI 3.6–10, p < 0.001 (Table 1). The patients with a positive bacterial pathogen detected had a cough significantly more often than those without a bacterial pathogen (Table 2). Both fever and rhinitis were more prevalent in the patients with a bacterial pathogen detected, varying according to the bacteria in question (Table 2). There was no difference in shortness of breath or chest pain between the groups.
| No bacterial pathogen detected, N = 782 | Difference 95% CI, p | Bacterial pathogen detected N = 130 | Haemophilus influenzae detected N = 63 | Streptococcus pneumoniae detected N = 55 | Atypical bacterial pathogen detected N = 17 | |
|---|---|---|---|---|---|---|
| Age, years, mean (SD) | 62.2 (17.1) | 6.8 (3.6–10), p < 0.0001 | 55.4 (18.4) | 58.9 (17.7) | 56.1 (17.6) | 41.2 (17.8) |
| Any respiratory symptom1 | 641 (82%) | −10% (−14% to −3%), p = 0.004 | 119 (92%) | 56 (89%) | 52 (95%) | 16 (94%) |
| Other possible infection symptom2 | 751 (96%) | 1% (−2% to 6%), p = 0.5 | 124 (95%) | 60 (95%) | 53 (96%) | 16 (94%) |
| Current smoker | 128 (16%) | 7% (−15% to 0.3%), p = 0.06 | 30 (23%) | 13 (21%) | 15 (27%) | 3 (18%) |
| Pneumonia3 | 102 (13%) | −22% (−30% to −14%), p < 0001 | 45 (35%) | 12 (19%) | 23 (42%) | 11(65%) |
| Antibiotic treatment | 463 (59%) | −20% (−27% to −12%), p < 0.0001 | 103 (79%) | 48 (76%) | 45 (82%) | 14 (82%) |
| CRP mg/L at entry, mean (SD) | 59 (76)4 | 51 mg/L (36–65 mg/L), p < 0.0001 | 110 (93)5 | 101 (92) | 121 (98)5 | 113 (89) |
| Admitted | 524 (67%) | 0% (−8% to 9%), p > 0.999 | 87 (67%) | 40 (63%) | 39 (71%) | 13 (76%) |
| Respiratory support | 273 (35%)6 | 7% (−2% to 15%), p = 0.09 | 36 (28%)5 | 16 (25%) | 17 (31%) | 6 (35%) |
| Oxygen | 233 (30%) | 4% (−4% to 12%), p = 0.3 | 33 (25%) | 15 (24%) | 15 (27%) | 6 (35%) |
| NIV | 24 (3.1%) | 1% (−4% to 3%), p = 0.6 | 3 (2.3%) | 1 (1.6%) | 2 (3.6%) | 0 |
| Ventilator | 16 (2.0%) | 2% (−1% to 3%), p = 0.08 | 0 | 0 | 0 | 0 |
| ICU care | 51 (6.5%) | 3% (−1% to 6%), p = 0.1 | 4 (3.1%) | 2 (3.2%) | 2 (3.6%) | 0 |
| Died | 27 (3.5%) | 2% (−2% to 4%), p = 0.3 | 2 (1.5%) | 2 (3.2%) | 0 | 0 |
- CRP, C-reactive protein; ICU intensive care unit; NIV, non-invasive ventilation.
- 1 Respiratory symptoms include cough, rhinitis, sore throat and shortness of breath.
- 2 Other possible infection symptoms include fever, chest pain and poor general condition.
- 3 Radiologically confirmed pneumonia.
- 4 Data for three patients lacking.
- 5 Data for one patient lacking.
- 6 Data for two patients lacking.
| No bacterial pathogen N = 782 n (%) | Haemophilus influenzae N = 63 n (%) | Proportion difference (95 CI) | p | Streptococcus pneumoniae N = 55 n (%) | Proportion difference (95 CI) | P | Atypical bacterial pathogen N = 17 n (%) | Proportion difference (95 CI) | p | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cough | 382 (49) | 44 (70) | −21 (−31 to −7.9) | 0.001 | 36 (65) | −17.4 (−29.2 to −3.7) | 0.01 | 15 (88) | −39.4 (−48.9 to −16.6) | 0.0006 |
| Rhinitis | 233 (30) | 30 (48) | −18 (−30 to −5.2) | 0.005 | 25 (45) | −15.4 (−28.8 to −2.5) | 0.02 | 5 (29) | 0.4 (−23.6 to 16.9) | 1 |
| Fever | 346 (44) | 34 (54) | −9.2 (−21.5 to 3.5) | 0.15 | 36 (65) | −23.2 (−34.8 to −9.3) | 0.001 | 13 (76) | −32.2 (−46.8 to −8.2) | 0.007 |
| Sore throat | 182 (23) | 26 (41) | −18 (−30.5 to −6.1) | 0.002 | 17 (31) | 8.0 (−21.6 to 3.3) | 0.14 | 4 (24) | −0.3 (−24.2 to 14.2) | 1 |
| Fatigue | 655 (84) | 53 (84) | 0.3 (−7.3 to 11.5) | 0.999 | 50 (91) | 8.2 (−13.6 to 2.2) | 0.08 | 16 (94) | −10.4 (−16.2 to 10.9) | 0.33 |
- There were less than 10 patients with data on symptoms lacking. These were counted as symptomless. There was no difference in shortness of breath or chest pain between the groups.
Either chest X-ray or thoracic CT scan was performed for 94% (856/912) of the patients. Pneumonia was diagnosed radiologically in 13% of the patients without any bacterial pathogen finding and in 35% of those with positive bacterial results (difference 22%, 95% CI 14% to 30%, p < 0.001, Table 1). Pneumonia was most common in patients with an atypical bacterial pathogen (11/17, 65%), although 42% (23/55) of those with positive S. pneumoniae detection also had pneumonia. The patients with a positive bacterial pathogen detection were significantly more likely to receive bacterial antibiotics (103/130, 79%) than those with negative results (463/782, 59%, difference 20%, 95% CI 12% to 27%, p < 0.0001).
A total of 611 patients of the 912 participants (67%) were admitted to the hospital. The mean C-reactive protein (CRP) concentration at the entry was higher in patients with a positive bacterial finding (110 mg/L vs 59 mg/L, the difference between the means being 51 mg/L, 95% CI 36–65 mg/L, p < 0.0001, Table 1). Intensive care was required for 6.5% (51/782) of the patients without any bacterial pathogen, compared with the value of 3.1% (4/130) for the patients with a positive result.
The multivariate model showed positive detection of S. pneumoniae to be associated with a 4.5-fold risk of pneumonia (Table 3) and positive detection of either C. pneumoniae or M. pneumoniae to be associated with an almost 9-fold risk (Table 3). The detection of S. pneumoniae was also associated with a history of chronic lung disease and a high CRP value upon arrival (Table 4).
| Risk factors for pneumonia | Adjusted OR2 (CI 95%) | p Value |
|---|---|---|
| Positive detection for an atypical bacterial pathogen1 | 8.96 (2.99–26.89) | <0.001 |
| Positive detection for Streptococcus pneumoniae | 4.49 (2.42–8.33) | <0.001 |
| Positive detection for Haemophilus influenzae | 1.34 (0.66–2.69) | 0.417 |
- 1 Atypical bacterial pathogen = Chlamydia pneumoniae or Mycoplasma pneumoniae.
- 2 Adjusted for age and gender.
| Variable | Adjusted OR1 (CI 95%) | p Value |
|---|---|---|
| CRP at entry, 10 mg/L | 1.06 (1.02–1.09) | 0.001 |
| History of chronic lung disease | 1.96 (1.07–3.58) | 0.029 |
| Cough at entry | 1.43 (0.77–2.65) | 0.262 |
| Fever at entry | 1.30 (0.62–2.70) | 0.485 |
- 1 Adjusted for age and gender.
We also evaluated the information obtained using other bacterial detection methods, adopted in routine care. Blood cultures were taken from 495/912 patients (54.3%) and 23/495 (4.6%) were found to be positive. Using this method, S. pneumoniae was found in seven patients, and the same pathogen was detected using PCR in four of these cases, but the result was negative in three. Five of the patients with S. pneumoniae in blood cultures had definitive pneumonia in radiological evaluation, and two of them were suspected to have pneumonia. Pneumococcal antigen was detected in the urine of 6 out of the 101 patients tested, and one of these patients had a nasal swab that was positive for S. pneumoniae in multiplex PCR.
DISCUSSION
This cohort of acutely ill adults enrolled in a hospital emergency room enabled us to show that bacterial PCR performed on nasopharyngeal samples contributed valuable information for the diagnosis of infections. A positive finding of S. pneumoniae was associated with a 4.5-fold risk of pneumonia and the detection of atypical bacteria with a 9-fold risk.
In the management of respiratory tract infections, rapid, accurate diagnosis of microbial aetiology is important for choosing the right treatment, whether antibiotics, antiviral medication or symptomatic care [10]. The multiplex PCR method has been used previously to detect viral respiratory pathogens, but only limited research has been carried out into nasopharyngeal bacterial pathogens. There is an increasing interest in wider PCR panels for the detection of bacterial pathogens in samples obtained using non-invasive methods that are suitable for outpatients as well [11, 12]. We evaluated here the degree to which bacterial pathogen detection results could reflect clinical features such as symptoms, radiological findings and other laboratory results taking the radiologist's decision on thoracic imaging, either a chest X-ray or a thoracic CT scan, as the criterion for pneumonia. According to the results, radiological pneumonia was associated with bacterial detections in PCR analyses of nasopharyngeal samples.
We found that patients with positive bacterial detection results were significantly more likely to have a cough. On the other hand, rhinitis and a sore throat were significantly more common in those detected positive for Haemophilus and fever in those detected positive for an atypical respiratory pathogen. Although it has been noted earlier in a meta-analysis of atypical respiratory pathogens that individual signs and symptoms alone are of little value for the diagnosis of these pathogens [13], the present study, using a multivariate analysis, showed that a positive detection of S. pneumoniae was significantly associated with a history of chronic lung disease and a higher concentration of C-reactive protein but not with any particular symptom.
Chronic lung disease has been noted as a risk factor in studies of pneumococcal disease [14, 15], and smoking has been recognized as such in several studies [16] and is thought to increase the prevalence of bacterial pathogens in the upper airways [17]. In the present cohort, smoking was more common among the patients with positive bacterial findings than among those with negative bacterial results, but the number of smokers was too low for any meaningful analysis.
It has been shown that the proportion of positive PCR detections decreases with age [18, 19], and this was also the case in this study. Since patients in internal medicine clinics are elderly, this may weaken the value of the diagnostic procedure involved. However, in a study by Azadeh et al., the pathogens found from nasopharynx and bronchoalveolar lavage (BAL) fluid were well in accordance [20]. The yield of positive detections was higher in BAL, but the method is not suitable for use in an emergency room or for all patients. The interpretation of the results achieved using different sampling methods is not yet clear. For instance, the use of new biomarkers is currently being investigated for better recognition of infection [21, 22]. Furthermore, our cohort was not only limited to patients with respiratory infection symptoms but also included some with chest pain or poor general condition, which presumably also reduced the proportion of positive results.
In particular, the role of positive detections of S. pneumoniae in respiratory samples was a highly controversial matter at the time when this cohort was formed, with the carriage rate in asymptomatic elderly patients being in the range of 0–5% when assessed using conventional culture-based methods [23-25], whereas it has been up to 22% at several sampling sites with PCR-based methods [26]. The pneumococcal colonization rate of children has been reported to be up to 10 times higher than that in adults [27]. The patients with positive bacterial findings in our cohort were younger, more often had pneumonia, more often received bacterial antibiotics and had higher concentrations of C-reactive protein at the primary evaluation. They also had more instances of cough and fever.
The strengths of our study are its large cohort and vast body of prospectively collected clinical data, including symptoms. The limitations are the lack of systematic data on other bacteriological diagnostic methods, such as blood cultures on all patients or cultures of the swab samples. Due to the observational study design, it is not possible to evaluate the impact of the bacterial findings on clinical decision making.
Nasopharyngeal samples may include pathogenic respiratory bacteria that are of no clinical importance. More than half of all children in day care may have H. influenzae or S. pneumoniae in their nasopharynx without any clinical signs of bacterial illness [28], but we were able here to show that observations of the pathogenic respiratory bacteria achieved with the PCR technique predicted the probability of pneumonia among adult patients. We suggest that bacterial PCR performed on nasopharyngeal samples can contribute valuable information for the diagnosis of infections. It was especially valuable here in predicting the occurrence of pneumonia among patients with Chlamydia pneumoniae or Mycoplasma pneumoniae infections. Nevertheless, we would emphasize that not all patients with positive bacterial samples require antimicrobial therapy and that the results should always be interpreted as part of the clinical entity.
We thank Nina Hannu for her help with enrolment of the patients and the collecting of data.
FUNDING
This was an academic investigator-driven study supported by the Research Foundation of the Pulmonary Diseases in English and Hengityssairauksien tutkimussäätiö in Finnish by funding the scientific work of ES with a grant. The Northern Ostrobothnia Hospital District, Finland, contributed to the laboratory costs of the study. The manufacturer of the multiplex PCR did not fund the work or participate in it in any other ways.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare that are relevant to the content of this article.
AUTHOR CONTRIBUTIONS
MR, MU, HK and TSR contributed to the study conception and design. TP was expert and consultant in data analyses. ES collected the data and drafted the manuscript. All authors critically reviewed and approved the final manuscript.
ETHICAL APPROVAL
The protocol was found ethically acceptable by the Ethics Committee of the Northern Ostrobothnia Hospital District, Oulu, Finland (EETTMK 45/2014 §137).
INFORMED CONSENT
Informed consent was obtained from all individual participants included in the study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on a reasonable request from the corresponding author, [ES], for clinical research purposes. The data are not publicly available as they contain information that could compromise the privacy of the research participants. Only data that do not compromise the privacy of the research participants will be shared.




