Distinct roles of types 1 and 2 interferons in human eosinophil regulation: A multi-omics analysis
Interferons (IFNs) are broadly categorised into types 1, 2, and 3, represented by IFN-α/β, −γ, and -λ, respectively.1 IFNs play pivotal roles in the immune response against infectious and malignant diseases. However, the roles of IFNs in the pathogenesis of allergic diseases that mainly exhibit type 2 cytokines-induced eosinophilic inflammation remain fully uncovered. Eosinophils are pivotal in allergic diseases with various cellular functions, including degranulation, cytokine release, reactive oxygen species production, and eosinophil ETosis at inflammatory sites.2 Herein, we investigated the regulatory effects of IFNs on human eosinophils.
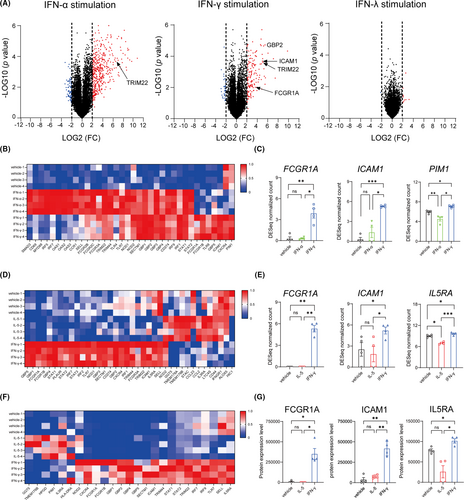
Multi-omics analysis comprising transcriptomics and proteomics using the cells isolated from healthy participants identified 19788 protein-coding genes and 6662 proteins in IFN-stimulated eosinophils. Volcano plots constructed to visualise expression signatures of the genes induced by IFN stimulation revealed alterations in the expression of the genes in IFN-α- and IFN-γ-stimulated eosinophils but not in those stimulated with IFN-λ (Figure 1A). Principal component analysis of factors significantly changed with IFN-α and IFN-γ stimulation showed distinct clustering (Figure S1A,B). IFN-α and IFN-γ upregulated anti-viral molecules including guanylate binding and tripartite motif-containing proteins (GBPs and TRIMs)3 (Figure 1B; Figure S1C). The expression GBP2 and TRIM22 were selectively upregulated by both IFNs; however, the changes in their expression were more pronounced in eosinophils stimulated with IFN-γ and IFN-α, respectively, (Figure S1D) and partly counterregulated by IL-5 (Figure S1E). In addition, IFN-γ, but not IFN-α, upregulated Fc gamma receptor-1A (FCGR1A), intercellular adhesion molecule 1 (ICAM1: a primary receptor for rhinovirus), C-X-C chemokine receptor type 4 (CXCR4), and PIM1, a pro-survival factor in human eosinophils (Figure 1C; Figure S1F).4, 5 Flow cytometric analysis showed the IFN-γ-induced expressions of FCGR1, ICAM1, and CXCR4 on cell surfaces (Figure S1G). Additional multi-omics analysis of eosinophils upon long-term stimulation with IFN-γ and IL-5 revealed that IFN-γ upregulaed the expressions of these molecules (FCGR1, ICAM1, and CXCR4) and IL5RA at both gene and protein levels, which pattern was clearly distinct from IL-5-induced one (transcriptomics: Figure 1D,E; Figure S2A–D; proteomics: Figure 1F,G; Figure S2E–H).
Mechanistic investigation using flow cytometry and quantitative real-time polymerase chain reaction showed the different effects of IFN-α and IFN-γ on cellular survival (Figure 2A,B). IFN-α inhibited IL-5-induced prosurvival effect (Figure 2A) and IFN-γ exerted PIM-1-dependent prosurivival property (Figure 2B). In addition, long-term stimulation with IFN-γ selectively upregulated (Figure 2C,D) and restored IL5RA expression that was downregulated upon IL-5 stimulation (Figure 2E–G). IFN-γ-induced upregulation of IL5RA enhanced eosinophil responsiveness to IL-5 with more prominent prosurvival effects (Figure 2H). Graphical explanations of these assays were made in Figure S3.
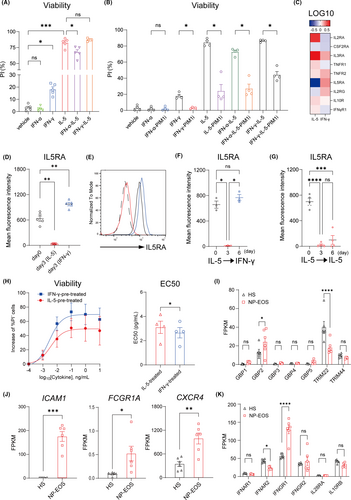
To investigate the pathogenic roles of IFN-related signalling in eosinophils of patients with allergic disease, previously reported RNA-sequencing data of eosinophils derived from nasal polyps (NP-EOS) in patients with chronic rhinosinusitis (N = 6) and healthy subjects (HP) (N = 6)6 on IFNs-related factors was evaluated. This analysis demonstrated that the expression pattern of IFN-related molecules in NP-EOS showed an augmented or diminished signal of IFN-γ (GBP2, FCGR1A, ICAM1, and CXCR4) or IFN-α (TRIM22) with the upregulated or downregulated expression of each receptor (IFNGR1 and IFNAR2) (Figure 2I,K). No differences were observed in the expression of IFN-λ receptors, IL28RA and IL10RB (Figure 2K). These findings suggest the possibility of enhanced IFN-γ- and attenuated IFN-α-signaling in NP-EOS.
In conclusion, the present study elucidated the mechanism of IFN-induced anti-viral properties and revealed that IFN-γ and IFN-α play distinct roles in the regulation of eosinophils. Multi-omics analysis using clinical specimens confirmed the dual effects of IFN-γ and IFN-α with pathogenic and protective significance. This approach demonstrated its methodological usefulness in identifying type 2 inflammation-independent mechanisms in refractory allergic diseases. IFN-γ specifically induced the expression of pro-inflammatory molecules like FCGR1A, ICAM1, PIM1, and IL5RA, suggesting that IFN-γ might contribute to the initiation and maintenance of chronic eosinophilic inflammation. Developing therapeutic strategies to selectively control type 1 and type 2 IFNs-signaling could be an innovative approach to treat refractory eosinophilic diseases.
AUTHOR CONTRIBUTIONS
J.M. and H.S. designed the experiments; H.S. and J.M. performed most of the experiments and compiled the data; Y.K. (Yusuke Kawashima) and Y.H. performed omics analysis; Y.K. (Yoshifumi Kimizuka), A.K., M.A., and K.F. supervised the project; H.S. and J.M. wrote the manuscript; R.O., Y.O., E.M., K.S., K.M., H.K., S.U., and K.A. discussed the results; and all authors provided feedback on the manuscript.
ACKNOWLEDGMENTS
We thank Tomoya Sano (National Defense Medical College), Ryohei Suematsu (National Defense Medical College), Yohei Maki (National Defense Medical College), and Takunori Ogawa (National Defense Medical College) for technical support to collect clinical sampled regarding this study.
FUNDING INFORMATION
This work was supported by Research Grant on Allergic Disease and Immunology from the Japan Agency for Medical Research and Development (24ek0410097; to J.M., S.U., and K.A.), JSPS Grant-in-Aid for Young Scientists (20 K17239; to J.M.), JSPS Grant-in-Aid for Scientific Research (C) (24 K11605 to J.M.), Asthma Basic Research Supporting Program of the Japan Asthma Society (to J.M.), RIKEN Special Postdoctoral Researchers Program (to J.M.), GSK Japan Research Grant 2018 (to J.M.), Grant-in-Aid for Research of the ONO Medical Research Foundation (to J.M.), JST-ERATO grant number JPMJER2101 (to M.A.), and JSPS Grant-in-Aid for Scientific Research on Innovative Areas (15H05897, 15H05898, 20H00495 to M.A.).
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The RNA-seq data generated in this study are available in the DDBJ (DNA Data Bank of Japan: https://www.ddbj.nig.ac.jp/index-e.html) database under BioProject PRJDB18153 and PRJDB18155 with GEA (Genomic Expression Archive) E-GEAD-806 and E-GEAD-807, respectively. Proteomic data used in this study has been deposited to the ProteomeXchange Consortium via the jPOST partner repository (http://www.proteomexchange.org/) with the dataset identifier PXD052615. Further inquiries can be directed to the corresponding author.