Advances and highlights in biomarkers of allergic diseases
Abstract
During the past years, there has been a global outbreak of allergic diseases, presenting a considerable medical and socioeconomical burden. A large fraction of allergic diseases is characterized by a type 2 immune response involving Th2 cells, type 2 innate lymphoid cells, eosinophils, mast cells, and M2 macrophages. Biomarkers are valuable parameters for precision medicine as they provide information on the disease endotypes, clusters, precision diagnoses, identification of therapeutic targets, and monitoring of treatment efficacies. The availability of powerful omics technologies, together with integrated data analysis and network-based approaches can help the identification of clinically useful biomarkers. These biomarkers need to be accurately quantified using robust and reproducible methods, such as reliable and point-of-care systems. Ideally, samples should be collected using quick, cost-efficient and noninvasive methods. In recent years, a plethora of research has been directed toward finding novel biomarkers of allergic diseases. Promising biomarkers of type 2 allergic diseases include sputum eosinophils, serum periostin and exhaled nitric oxide. Several other biomarkers, such as pro-inflammatory mediators, miRNAs, eicosanoid molecules, epithelial barrier integrity, and microbiota changes are useful for diagnosis and monitoring of allergic diseases and can be quantified in serum, body fluids and exhaled air. Herein, we review recent studies on biomarkers for the diagnosis and treatment of asthma, chronic urticaria, atopic dermatitis, allergic rhinitis, chronic rhinosinusitis, food allergies, anaphylaxis, drug hypersensitivity and allergen immunotherapy. In addition, we discuss COVID-19 and allergic diseases within the perspective of biomarkers and recommendations on the management of allergic and asthmatic patients during the COVID-19 pandemic.
1 INTRODUCTION
Allergic diseases represent a collection of disorders mediated by innate and adaptive immune responses along with epithelial cells that cooperate and cause immunologic hypersensitivity toward non-self and otherwise harmless environmental molecules. Allergic diseases including asthma, atopic dermatitis (AD), allergic rhinitis (AR), conjunctivitis, chronic rhinosinusitis (CRS) and food allergy affect over 25% of the population in industrialized countries and are increasing in prevalence in developing countries representing a significant socioeconomic burden. Several factors including pollution, climate change, reduction in biodiversity, urbanization, change in lifestyle and dietary habits have been attributed to the important rise in allergic cases.1-4
Precision medicine, a customized treatment approach, and lifestyle changes can reduce the socioeconomic burden imposed by allergic diseases. These are particularly suited for precision medicine as the representing umbrella characteristics, such as type 2 immunity, comprise different diseases linking an underlying pathomechanism to an endotype or phenotype. The widespread use of precision medicine warrants the discovery of novel biomarkers as measurable indicators of physio pathological conditions. These biomarkers can be used as diagnostic tools, to monitor disease progression, to select the most effective therapy, and to aid in the prognosis of treatment outcome.5, 6
Biomarkers are indicators of normal biological processes, pathogenic processes or biological responses to therapeutic intervention that are objectively measured. Asthma and other allergic diseases are complex disorders with several disease variants due to different underlying cellular and molecular mechanisms. A biomarker for allergen-specific immunotherapy will rather likely be involved in the modification of an immune parameter linked to inflammation. Several biomarkers have been reported for allergic diseases and asthma (eg, IgE, blood or sputum eosinophilia, fractional exhaled nitric oxide [FeNO], serum interleukin (IL)-5 and IL-13) or are under current investigation (eg, pro-inflammatory mediators, genes, microRNAs (miRNAs), markers of epithelial barrier integrity, microbiomes).7, 8 In this review, we summarize the recently reported biomarkers for allergic diseases and further discuss them in the context of COVID-19.
2 BIOMARKERS IN ASTHMA
2.1 Biomarkers related to endotypes and phenotypes of asthma
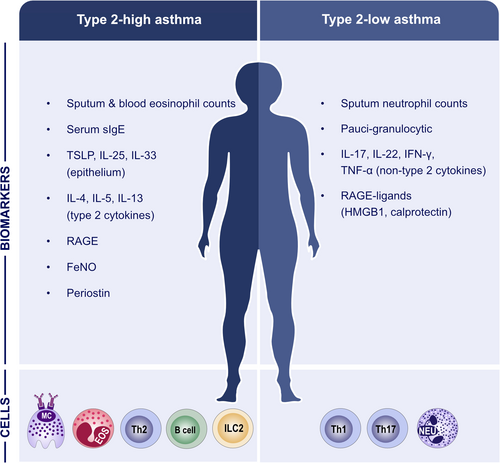
The heterogeneity of asthma with several phenotypes and endotypes makes it particularly suited for precision medicine, and the identification of different noninvasive biomarkers will facilitate its diagnosis and treatment. Two main subtypes of immune responses driving asthma have been defined, namely type 2-high and type 2-low.1, 9, 10 A specific type 2 set of cytokines is produced during the induction and maintenance of an allergic immune response with the contribution of epithelial cells, dendritic cells (DCs), innate lymphoid cells (ILCs), T cells, eosinophils, mast cells and basophils. Activation of Th2- and ILC2-pathways are at the core of type 2 inflammation in asthma. Th2 cytokines include IL-4, IL-5, IL-9, IL-13, IL-31, whereas the main type 2 cytokines produced by ILC2s are IL-5, IL-9 and IL-13.11 Progress has been made in profiling type 2 immune response-driven diseases, however there are limited studies of non-type 2 immune responses in asthma, CRS and AD. Eosinophils, IgE, circulating chemokine receptor (CCR) 10 expressing ILC2s, plasma CCL27 levels, and FeNO are common well-known biomarkers that have been reported for the diagnosis and prognosis of asthma. In addition, immunoglobulin E (IgE), subclasses of IgG, serum inhibitory activity for IgE, basophil activation, chemokines and cytokines, regulatory T (Treg), Breg, and DC markers have been suggested as useful markers to follow allergen tolerance-inducing treatments. Moreover, an electronic nose and a mobile airways sentinel network have been developed as promising biomarker tools to improve our understanding of the phenotypical characteristics of asthma and the multimorbidities between different allergic diseases.12
The characterization of genetic variations between non-eosinophilic and eosinophilic asthma has been performed through transcriptomics studies.13 Cluster analyses of the differentially expressed transcripts have revealed three distinct clusters: the highly eosinophilic type cluster is characterized by the transcript expression pattern of IL-33R, CCR3, and thymic stromal lymphopoietin (TSLP) receptor (TSLPR), and shows gene signatures associated with type 2-high mechanisms/pathways; patients showing this transcript pattern had the highest sputum eosinophilia and exhaled nitric oxide fraction, and had severe asthma; the highly neutrophilic type cluster showed the highest sputum neutrophilia, serum C-reactive protein levels and prevalence of eczema and was characterized by transcript expression patterns of interferon (IFN) and tumor necrosis factor (TNF) superfamily genes; the paucigranulocytic-eosinophilic type cluster is characterized by genes of metabolic pathways, ubiquitination and mitochondrial function and shows the lowest prevalence of severe asthma.14, 15 McDowell and Heaney evaluated sputum and blood eosinophil counts and FeNO as useful biomarkers of type 2-high asthma. Clinical assessments indicate that type 2-low patients present multiple non-asthma extra-pulmonary manifestations.16 Type 2-high asthma and allergic airway disease development have been reported to be substantially controlled by the receptor for advanced glycation endproducts (RAGE) through type 2 inflammatory response. In contrast, elevated levels of the RAGE ligands, high-mobility group box-1 and calprotectin, were significantly associated with neutrophilic asthma severity, exacerbation risk, and steroid-insensitivity (Figure 1).17 Regarding IL-1 and TNF-α/NF-κB pathways, upregulation of IRAK3 and NFBIZ transcript expression in peripheral blood neutrophils and IRAK3, IRAK2, and IL1R2 in airways have been suggested as indicators of transcriptional asthma phenotypes which are characterized by increased neutrophils.13
Endo et al. suggested that high FeNO levels may be indicative of poor short-term lung function, as measured by changes in oscillometric parameters. In this respect, deterioration of respiratory system resistance at 5 Hz (R5), the difference between respiratory system resistance at 5 Hz and 20 Hz (R5–R20), respiratory system reactance at 5 Hz, resonant frequency, and area under the reactance after a 3-month period was observed in the high FeNO group.18 Baseline FeNO levels in the high FeNO group were positively correlated with changes in R5, R20, and R5–R20. However, this observation was not substantiated in respiratory system reactance at 5 Hz, resonant frequency, and area under the reactance measurements. Moreover, a negative association between FeNO levels and R5 and R20 was found to be significant. This study suggests higher baseline levels of FeNO as a potential biomarker of changes in lung resistance, but not reactance.18
The chromosome 17q21 is of interest in genetic epidemiological studies on asthma as it contains crucial genes related to its pathology, including ORMDL3, GSDMB, LRRC3C, GSDMA, ZPBP2, IKZF3, GRB7, ERBB2, and PGAP3. In addition, three SNPs (rs4794820, rs807631, rs2872507) that are located in this chromosome were significantly associated with the pathogenesis and severity of the disease.19
Identifying treatable traits may provide a stratified approach for improving the physiological symptoms and mental well-being of asthma patients.20 Treatable traits in asthma can be classified as pulmonary, extra-pulmonary and behavioral/psychosocial traits. Pulmonary treatable traits are fixed airflow limitation, bronchodilator reversibility, type 2 and neutrophilic inflammation, cough, exercise-induced respiratory symptoms, and bronchitis. The most prevalent extra-pulmonary traits are rhinosinusitis, obesity, obstructive sleep apnea, reflux, and atopy. Behavioral/psychosocial traits include smoking, poor adherence to medication, anxiety and other psychiatric disorders.20 There is a pressing need for control of environmental hazards to ameliorate the development of asthma.21
2.2 Biomarkers for treatment prognosis with biological drugs
Allergic diseases including severe asthma impose a burden on patients and healthcare system. Due to the less effectiveness of nonbiological add-on treatments and adverse effects of repeated oral corticosteroid courses, the need for the development of novel therapies has been crucial. Recently, new therapeutic options have become available referred to biologicals, which are mainly genetically synthesized monoclonal antibodies (mAb) or other proteins with a high-molecular weight that are used to diagnose, prevent and treat different diseases. Among them, mAb (Table 1) can selectively bind and inhibit a specific biological determinant that makes them ideal for use in personalized or precision medicine.
| Biological | Description and target | Add-on treatment | Benefits | Prediction of response |
|---|---|---|---|---|
| Dupilumab |
Anti-IL-4/IL-13 (IL-4Rα) Human mAB against IL-4 receptor Ra |
Uncontrolled severe eosinophilic asthma in adults and adolescents aged 12 years and older |
Reduce systemic steroid use Reduce the rate of severe exacerbations Improve lung function |
Higher baseline levels of eosinophils ≥150 cells/μl26 FeNO ≥25 ppb26 |
| Omalizumab |
Anti-IgE Humanized mAB against IgE |
Uncontrolled severe allergic asthma in adults, adolescents and children aged 6 years and older |
Reduction in severe exacerbations Reduce OCS use Safe to continue during pregnancy |
Independent of blood eosinophil cutoff23 (either <or ≥300 cells/µl)25 Blood eosinophils <300 cells/µl FeNO <50 ppd26 Blood eosinophils ≥300 cells/μl23, 25, 26 and IgE >75 IU/ml25 Specific eosinophil (≥260/μl) and FeNO (≥19.5 ppb) cutoffs23 |
| Benralizumab | Anti-IL-5 receptor Humanized mAB against IL-5 receptor Ra | Uncontrolled severe eosinophilic asthma in adults and adolescents aged 12 years and older |
Reduce OCS use Reduction in severe exacerbations |
High blood eosinophil counts ≥300 cells/μ.26 |
| Mepolizumab |
Anti-IL-5 Humanized mAb against IL-5 |
Uncontrolled severe eosinophilic asthma in adults, adolescents and children aged 6 years and older |
Decreases total use of OCS Reduction in exacerbations |
Higher blood eosinophil counts ≥150–300 cells/μl24 High blood eosinophils ≥150 cells/μl26 |
| Reslizumab |
Anti-IL-5 Humanized mAb againstIL-5 |
Uncontrolled severe eosinophilic asthma in adults aged 18 years and older |
Reduce OCS use Reduction in exacerbations |
High blood eosinophil count >400 cells/μl26 |
- Abbreviations: FeNO, fractional exhaled NO; IgE, immunoglobulin E; IL-13, interleukin-13; IL-4, interleukin-4; IL-4Rα, interleukin-4 receptor-alpha; IL-5, interleukin-5; OCS, oral corticosteroid.
A European Academy of Allergy and Clinical Immunology (EAACI) position paper broadly reviewed the currently approved biologicals and small molecule drugs under clinical research for the treatment of asthma.22 Roth-Walter et al. highlighted the challenge of patient stratification due to a lack of small chemical drug-related biomarkers and an unrevealed broader spectrum of biological functions induced by small molecules. The authors emphasized the need of including multi-omics stratification tools to overcome the complexity of adequate biomarker identification.22
The EAACI Guidelines on the use of biologicals in severe asthma highlighted a conditional recommendation of using dupilumab, a human monoclonal antibody binding to the alpha subunit of the IL-4 receptor, as an add-on treatment for adult patients with severe eosinophilic asthma and those who have severe corticosteroid-dependent asthma irrespective of blood eosinophil levels (Table 1). In adult patients with severe asthma, a ≥150/μl blood eosinophils cutoff point was recommended to identify patients with a good likelihood for response to anti-IL-5 therapy. Anti-IgE therapy was suggested for adolescents and adults with cutoffs of specific eosinophil counts ≥260/μl and FeNO levels ≥19.5 ppb.23
Numerous mAbs that target mediators associated with type 2 high inflammation are currently available for treating severe asthma. Blood eosinophil counts, the biomarker of choice in asthma management, can be used to discriminate between type 2 and non-type 2 asthma. However, blood eosinophil counts are of limited value for precision medicine as they cannot be further sub-grouped and so there is a pressing need to identify biomarkers of the dominant inflammatory mediator driving disease pathogenesis in the organ of interest. A recent editorial published in 2019 emphasized the importance of assessing the “rate of exacerbations”, which was suggested to be a more objective outcome to evaluate the optimal therapeutic effect of mepolizumab in severe eosinophilic asthma patients, as a subgroup of patients remain symptomatic despite normalization of blood eosinophil levels after treatment.24
A reduction in oral corticosteroid use is the outcome of a significant reduction in the annual rate of severe exacerbations and is crucial for patients. In this regard, Papadopoulos et al.25 highlighted the ease of FeNO measurements as a biomarker of the efficiency of dupilumab.
Eyerich et al. reviewed recent and upcoming developments in the treatment of allergic diseases that emphasized the importance of predicting an enhanced response to the three IL-5-targeting approaches (mepolizumab, reslizumab, benralizumab), all of which show a similar reduction in asthma exacerbations. Blood eosinophil levels, co-existence of nasal polyps and body weight were suggested as predictive parameters for biological treatment success in severe asthma.26
Implementing biologics into the care pathways for CRS with nasal polyp (CRSwNP) patients with and without comorbid asthma has been recently proposed. Since anti-IgE, anti-IL5, and anti-IL4Rα biologics were demonstrated to have a beneficial effect in patients with comorbid asthma, they were recommended for CRSwNP patients with a type 2 inflammatory profile.27, 28
Carlsson et al. investigated prognostic biomarkers for the clinical response to azithromycin treatment in young children with asthma-like symptoms. This prospective study observed alterations in the upper airway immune mediator levels during episodes of asthma-like symptoms and suggested that levels of TNF-α, CCL22, and IL-10 may predict the response to azithromycin treatment.29
Blocking the colony stimulating factor 1 receptor (CSF1R) on circulating DCs selectively inhibits the production of Th2 memory cells. The findings reported by Moon et al.30 showed that inhibiting the CSF1-CSF1R pathway ameliorates bronchial hyperresponsiveness in a murine chronic asthma model.
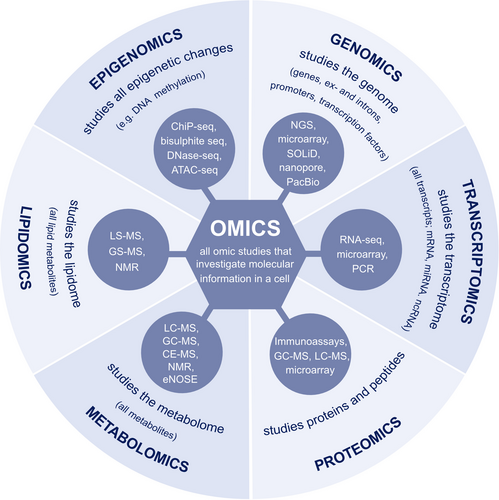
2.3 Omics and biomarkers in asthma
As our understanding of asthma complexity improves, it becomes increasingly evident that the heterogeneity in its underlying pathophysiology and treatment response demands for a proactive personalized approach.7, 31 The Human Genome Project launched in 1990 gave way to an exponential growth of open databases that fueled the progress of omics technologies that have recently become economically viable, facilitating research of diseases at a molecular level.32 The omics technologies – epigenomics, genomics, transcriptomics, proteomics, metabolomics, lipidomics (Figure 2) – focus on the identification of molecular biomarkers to facilitate clinical decision-making in the future.31, 32
The genomics approach aims to identify gene variants associated with specific phenotypes mainly through candidate-gene studies and genome-wide association studies (GWAS). A systemic review over several GWAS studies estimated a 10%–29% positive response to short acting beta-agonists for patients with certain genes variants, including variants of the beta-2 adrenergic receptor gene ADRB2. However, asthma susceptibility is difficult to link to just one or several genome variations due to the complexity of the disease.32, 33 Further large-scale approaches are warranted and are starting to emerge, such as those conducted in the U-BIOPRED consortium and the Severe Asthma Respiratory Program.34
The U-BIOPRED consortium performed transcriptomic analysis from a large asthma cohort and found over 1500 differentially expressed genes in asthmatics compared to non-asthmatics.32 Transcriptomic technologies comprise real-time PCR, microarrays and RNA sequencing. Micro RNAs (miRNAs) have received increasing attention during the last decade. These are short noncoding RNAs that act as post-transcriptional negative regulators by binding to mRNA, which leads to their cleavage and degredation.35, 36 Hirai et al.36 characterized the asthma-COPD overlap by using a qPCR array that identified five miRNAs as potential biomarkers to discriminate asthma-COPD overlap patients from those suffering from asthma alone. The ability for miRNAs to target several mRNAs suggests that they play an important role for miRNAs in the pathophysiology of various diseases, and their use as potential biomarkers and therapeutic targets are just beginning to emerge.35 Transcriptomics analyses of airway smooth muscle from asthmatics revealed several differentially expressed genes that correlated with airway hyperresponsiveness (AHR). Interestingly, corticosteroid-induced expressional changes of certain genes were associated with improved airway physiology.34 Hernandez-Pacheco et al.37 used RNA sequencing of various datasets to study response to inhaled corticosteroids (ICS) among asthmatics and found that LTBP1 is a potential biomarker for treatment response.
Transcriptomics approaches are continuously evolving and single-cell transcriptomics is currently routinely performed to gain insight into functional cell subsets. Jackson et al.38 investigated the whole-transcriptome responses to treatment of single human airway epithelium cells to IL-13. IL-13 was found to induce a transcriptional response and a functional transformation of epithelial cells, resulting in a metaplastic epithelium producing pathologic secretions and lacking innate airway defenses. Single-cell omics is also attracting attention in epigenomics studies of gene-environment interactions, where tissue and cell type context are of great importance.32
Both proteomics and metabolomics studies have been recently used as tools for the identification of novel biomarkers. Nieto-Fontarigo et al. optimized a bottom-up/nontargeted protocol to study human serum samples by means of proteomics. Combining liquid chromatography coupled online to mass spectrometry with the use of iTRAQ isobaric labels allowed them to identify 18 proteins as potential biomarkers of asthma phenotype and disease severity (eg, FCN2 and MASP1 for non-allergic asthma, or HSPG2 and IGFALS for allergic asthma).39 Abdel-Aziz et al. used an emerging metabolomic tool termed “breathomics” to successfully distinguish healthy from asthmatic subjects using an electronic nose. This method measures exhaled breath volatile organic compounds and its noninvasiveness is a key advantage for its use in the clinical practice.40
Omics technologies have greatly improved our understanding of the molecular mechanisms in asthma and play an important role in the discovery of biomarkers and targeted treatment options for type 2 asthma. Omics-based research is a valuable tool to further elucidate asthma complexity and to identify clinically applicable biomarkers. This is especially important for the development of treatment options for patients that are nonresponsive to currently available medications, including those suffering from non-type 2 asthma. Omics approaches in asthma research will undoubtedly continue to play a major role in progressing from a one-size-fits-all treatment approach to personalized management based on pathophysiological mechanisms and treatable traits.7, 32
2.4 Type 2 related biomarkers in asthma
There are currently five different biologicals available for the treatment of asthma, namely benralizumab, dupilumab, mepolizumab, omalizumab and reslizumab (Table 1). Allergic asthma is attributed to a Th2 immune response, and IL-4 and IL-13 are decisive cytokines for IgE class-switching and production, whereas IL-5 is responsible for peripheral and tissue eosinophilia. At the cellular level, the association between eosinophils and asthma is well-established.41 However, there is a continuous debate on whether blood or tissue eosinophil numbers are a more valuable parameter. Eosinophils in sputum are not solely related to allergic asthma, and measuring blood eosinophils to monitor therapeutic response provided limited information.41 It is important to determine the changes in laboratory parameters as early as possible to decide whether it is cost-effective to continue therapy for at least a year, or timely cease treatment and move to alternatives. Although there has been limited success in clinical practice at the cytokine level, anti-type 2 approaches have tremendous therapeutic potential.42 Serum IL-4 concentrations were correlated to the persistence of asthma in middle-age asthmatic patients and IL-6 was found to be a potential marker for early-onset asthma prognosis.43 Recently, Khumalo et al.42 found that the knockout of IL-4Rα, the common receptor subunit of IL-4 and IL-13, could significantly improve allergic asthma symptoms in mouse models. These results further confirmed the therapeutic potential of targeting the IL-4Rα targeting therapy. However, the levels of IL-33 receptor (ST2) were reduced in the bronchial epithelium of asthmatic patients compared to the control group. This trend was proposed to be induced by the pro-inflammatory environment, but raised doubts on the viability of targeting the ST2/IL-33 pathway.44 The number of circulating Th2 cells was found to be associated with the scores of the asthma control and of the asthma quality of life questionnaire scores during an asthma exacerbation. However, the percentage of CRTh2+ CD4+ T cells reversely correlated with lung function during the follow-up.45 Further research is warranted in using immune cells as biomarkers.
A study found that the number of IgE reactivities against respiratory allergen molecule is associated with asthma, rhinitis and conjunctivitis multimorbidity phenotypes compared to controls without any of these allergic diseases. Furthermore, the number of IgE reactivities were significantly increased for rhinoconjunctivitis multimorbidity as compared to rhinitis only or conjunctivitis only.46 Mas-related G protein-coupled receptor-X2 (MRGPRX2) is an endogenous receptor expressed in mast cells associated with IgE-independent activation and was recently found as a new biomarker for allergic asthma. Serum MRGPRX2 levels were elevated in allergic asthma patients, especially in those patients with good response to ICSs, which indicated that severe uncontrolled patients with lower levels of MRGPRX2 might require additional treatments.47 Protein S, a glycoprotein related to anti-inflammatory, anti-apoptotic, and anticoagulant processes has been recently found to play a role in the development of allergic asthma.48 The level of protein S in circulation was higher in asthmatic patients compared to the control group. Functional experiments in mice suggested that protein S could aggravate AHR and promote skewing toward a Th1 inflammatory response.48
Recently, the role of metabolites in allergic diseases has attracted research interest. Eicosanoids, comprising thromboxanes, leukotrienes, prostaglandins, and lipoxins, play a role in several pathophysiological processes in allergic diseases.49 In 2020, the EAACI Task Force published the first part of a consensus reached on the role of eicosanoids in the development of allergic diseases, clinical related findings and opportunities for further research. In the second part of the consensus, further disease links and pathophysiological questions were discussed.49
2.5 Biomarkers of the microbiome and epithelial barrier in asthma
There is a plethora of evidence indicating the important role of human microbiota in immune development and regulation.50 Poor microbial diversity during early-life has been suggested to contribute to the development and severity of immune-mediated diseases. In the last six decades, there has been an increase in allergic diseases correlating with urbanization, industrialization, Westernized lifestyle changes (eg, diet and obesity), increased rates of birth by cesarean delivery and increased early use of antibiotics.51, 52 These lifestyle changes are strongly affecting early-life microbial exposure, resulting in reduced gut microbial diversity. The association between human microbiota and allergic diseases is well-known.7, 50 Here, we summarize some of the recent key findings regarding the use of microbiota as a biomarker of asthma development, severity, and treatment follow-up. Epithelial changes associated with microbial dysbiosis, epithelial activation and epithelial barrier leakiness are suggested to be the culprit of many non-infectious inflammatory diseases as postulated in the “Epithelial Barrier Hypothesis” discussed in a comprehensive review.21 These processes not only underlie the development of allergy and autoimmune conditions in tissues with an impaired barrier function, but also apply to a wide range of diseases in which immune responses to translocated bacteria have systemic effects.21 The review article lists allergic diseases and autoimmune, metabolic disorders known to arise from local barrier defects and leakiness of the gut epithelium. Distant inflammatory responses due to a ‘leaky gut’ and microbiome changes are suspected to be associated with several neuropsychiatric diseases.21
Both airway and gut microbiota shape asthma pathogenesis, and asthma patients are characterized by a dysbiotic airway microbiome.7 A recent study compared the upper airway microbiota between asthmatic and non-asthmatic young adults and the elderly. Elevated levels of proteobacteria were found in asthmatics compared to non-asthmatic controls. Transcriptomic analysis of nasopharyngeal swabs from asthmatic patients indicated a higher relative abundance of microbiome genes related to the pentose phosphate pathway, lipopolysaccharide biosynthesis, flagellar assembly, and bacterial chemotaxis.53 A dysfunctional relationship between respiratory microbes and circulating metabolites in mite-sensitized childhood asthma was found using an integrated metagenomics-metabolomics approach. Total and mite allergen-specific IgE levels were associated with Streptococcus intermedius in children with mite-sensitized asthma. Moreover, the microbiome metabolic pathways could play a role in orchestrating inflammatory and allergic responses such as in biofilm formation by Pseudomonas aeruginosa, membrane trafficking and glycosaminoglycan degradation, all of which have been found to be over-activated in the asthmatic microbiome.54
In addition to the upper airway microbiota, a poor gut microbiota composition, especially in the first year of life, was associated with an increased risk of asthma development.7 At age 2–4, a high relative abundance of Gemmiger and Escherichia genera and low relative abundance of Collinsella and Dorea were predictive of subsequent development of asthma at age 6 years.55
Microbial products of healthy microbiota are also essential for maintaining human health. In recent years, short-chain fatty acids (SCFA) have been the most widely studied microbial gut metabolites. SCFA levels during the first year of life, especially butyrate, were associated with a decreased risk for asthma, food allergy, and allergic rhinitis. In addition, lower levels of butyrate metabolites were associated with the development of rhinitis and asthma in children. Moreover, total serum and mite allergen–specific IgE levels were associated with increased fecal butyrate levels in children with asthma.56 Fatty acids with immunomodulatory effects have been recently reported as important nutritional supplements for the prevention and treatment of asthma, especially during pregnancy and in infants. Metabolites derived from omega-3 or omega-6 (n−3, n−6) polyunsaturated fatty acids (PUFA) regulate inflammation. High levels of n−6 PUFA and n−3 PUFA were associated with exacerbated asthma symptoms and decreased risk of asthma, respectively, suggesting a protective effect of n−3 PUFA supplementation during pregnancy and infancy.57
The same environmental exposures that alter the human microbiota are also responsible for causing epithelial barrier dysfunction. As the first line of defense that provides a protective wall against environmental factors, impairment of this important structure disturbs homeostasis at mucosal sites and even in the affected organ, as demonstrated in many allergic, autoimmune and neuropsychiatric diseases.21, 58, 59
Wawrzyniak et al. found that CpG methylation of genes involved in the regulation of proteins with a role in maintaining bronchial barrier integrity was increased in asthmatic bronchial epithelial cells. Inhibition of DNA methyltransferase with SGI-1027 restored epithelial barrier integrity in vitro by increasing expression of tight junction proteins.60
In addition to barrier function, epithelial cells play other functional roles. One of the main characteristics of asthma is enhanced airway smooth muscle contraction and AHR. A recent report investigated the role of stanniocalcin-1 (STC1) on AHR and airway smooth muscle contraction. STC1 is an epithelial-derived protein whose levels in serum were positively correlated with asthma control and lung function. STC1 down-regulated stromal interaction molecule 1 and blocked store-operated Ca2+ entry resulting in a Ca2+ dependent suppression of smooth muscle contraction and therefore in airway smooth muscle relaxation. At the same time, IL-13 levels correlated negatively with serum STC1 levels in vivo and down-regulated STC1 expression in airway epithelial cells in vitro.61
2.6 Biomarkers of respiratory infections in asthma
Respiratory syncytial virus (RSV) and rhinovirus (RV) are the most common pathogens causing bronchiolitis in infants. Hospitalizations are associated with an increased subsequent risk of asthma in childhood. Tracheal aspirates from RSV-infected infants showed elevated levels of the pro-inflammatory cytokine IL-1β, which was induced by uric acid after RSV-infection. In a neonatal murine model of RSV-infection, the inhibition of IL-1β and uric acid reduced ILC2 infiltration to the lungs, which could further impede the type 2 immune response. This model suggested xanthine oxidase or the IL-1 receptor as new therapeutic targets to reduce asthma development related to RSV-infection.62 Furthermore, another study showed the association of increased levels of type 2 cytokines (IL-4, IL-5, IL-13, and TSLP) in the airways of infants with solo RV infection and the increased risk of developing childhood asthma. However, the levels of these cytokines were not different in asthmatic and non-asthmatic infants with RV/RSV coinfection.63 Early systemic anti-inflammatory intervention using corticosteroids could reduce the risk of subsequent RV-induced asthma by decreasing type 2 cytokines and the inflammatory response.64 Moreover, recurrent infections in severe asthma patients could be due to deficiency of local airway immune responses including specific antibodies. Ho et al. reported that primary Ig deficiency was not contributing to the recurrence of infections in severe asthmatics. However, administration of intravenous Ig in severe asthmatics increased sputum IgA and was associated with fewer infection-related exacerbations. Therefore, an uncompensated deficiency of IgA could cause recurrence of infections in asthmatics with recurrent infections.65
3 BIOMARKERS IN CHRONIC URTICARIA
Following oral aspirin challenge, the levels of pro-inflammatory urinary eicosanoid mediators increased in NSAID-induced urticaria/angioedema (NIUA) patients similar to those with NSAID-exacerbated cutaneous disease. This was attributed to the inhibition of cyclooxygenase 1 by NSAIDs, diverting the metabolism of arachidonic acid from prostaglandin E2 (PGE2) toward leukotriene E4 synthesis.66
In chronic inducible urticaria, patients often suffer from concurrent chronic spontaneous urticaria (CSU) and vice versa. It is important to identify triggers of chronic inducible urticaria based on patient history and provocation testing.67
Autoimmune CSU is immunologically distinct subtype of CSU and defined by the presence of mast cell-degranulating IgG autoantibodies to IgE or its FcεRI receptor. It is diagnostically confirmed by three criteria, namely a positive autologous serum skin test, basophil reactivity, and IgG anti-FcεRI/IgE. In a study by Schoepke et al., only 8% of the CSU cases fulfilled all three autoimmune CSU criteria. The immunological profile of autoimmune CSU patients was similar to basophil reactivity+ CSU patients but not autologous serum test+ or IgG anti-FcεRI/IgE+ CSU patients. Therefore, a positive basophil activation test (BAT) or basophil histamine release assay could be used to identify autoimmune CSU patients in clinical practice.68
4 BIOMARKERS IN ATOPIC DERMATITIS
Assessment of disease severity and understanding the underlying pathophysiological mechanisms behind the complexity of AD is warranted for personalized medicine, monitoring treatment effects, and clinical trials.69, 70
There is a great demand to develop minimally invasive approaches that allow for serial sampling of skin, especially for early-onset AD in infants and children. Pavel et al.71 reported that transcriptome profiling using tape strips in early-onset pediatric AD captures immune and barrier alterations in both lesional and nonlesional skin. Thijs et al. showed dried blood spot testing as a minimally invasive alternative to venipuncture. CCL17/thymus and activation-regulated chemokine (TARC), CCL18/pulmonary and activation-regulated chemokine, CCL22/macrophage-derived chemokine and I-309 levels measured in blood spots were strongly correlated with disease severity of moderate-to-severe AD and significantly decreased during effective treatment with topical steroids.72 In a recent review, Renert-Yuval et al.73 reported that other Th2 related chemokines including CCL26/eosinophil-attracting chemokine, CCL27/cutaneous T-cell–attracting chemokine, and Th2 related cytokine IL-13 and Th22 related cytokine IL-22 were also correlated with disease severity in untreated or post-treated tissues.
In healthy individuals, the skin microbiome is tightly regulated by intrinsic factors, but not in AD patients. The pH-dependent Staphylococcus aureus abundance is a predictor of AD severity.74 On the skin of children with AD in the MPAACH cohort, the presence of staphylococcal biofilms was confirmed. Strains of S. aureus showed higher relative biofilm propensity compared with S epidermidis, and were associated with increased severity of AD and increased transepidermal water loss.75 Interestingly, the microbiome and distinct disease-related gene expression profiles depend on anatomical location. Ottman et al.76 showed that the relative abundance of S. aureus is associated with disease severity in the posterior thigh, but not in the upper back.
The microbiota are also important as a therapeutic target. Boutin et al. identified nine bacterial genera whose abundance were inversely associated with the risk of developing allergic disease manifestations at 1 year of age. Microbiota profiling may be useful in clinical trials for the development of biotherapeutic products for the primary prevention of allergic disease and atopic multimorbidity.77
Several key studies on dupilumab in AD have been reported. A systematic review evaluated the efficacy, safety and economic impact of dupilumab in seven randomized controlled trials, which included in total 1845 moderate-to-severe AD patients treated with dupilumab.78 In both adults and adolescents, treatment with dupilumab significantly reduced short-term AD symptoms and severity, the use of rescue medications and improved quality of life. In addition, a long-term benefit was reported in adults. However, the cost-effectiveness ratio may be acceptable for high-income countries, but not in countries with limited resources.78
A study including 138 difficult-to-treat adult AD patients showed that dupilumab in daily practice improves disease severity, pain/discomfort, itch, anxiety, depression and health-related quality of life.79 Additionally, treatment with dupilumab significantly suppressed severity-related serum biomarkers such as TARC, pulmonary and activation-regulated chemokine, periostin, IL-22, and eosinophil-related markers such as eotaxin-1 and eotaxin-3.79 In patients without concomitant immunosuppressive therapy, dupilumab decreased the number of inflammatory cells and cytokines involved in AD pathogenesis in lesional skin (IL-5, IL-9, IL-13, IL-15, IL-17, IL-22, IFN-γ). As a consequence of the lower levels of type 2 cytokines in the skin, dupilumab also improved skin barrier function by increasing the expression of filaggrin, epidermal protease inhibitor lympho-epithelial Kazal-type-related inhibitor, and of the antimicrobial peptides human beta-defensin 3 and cathelicidin LL-37.80
Another systematic review clinically appraised 50 randomized controlled trials investigating the safety and efficacy of systemic treatments for AD in a total of 6681 patients. The most robust and high-quality trial evidence was found for the efficacy and safety of dupilumab up to 1 year in adults. An evidence-based robust clinical trial was also demonstrated for azathioprine, baricitinib and cyclosporine A. Assessment of the safety and efficacy is challenging due to considerable limitations in trial design, outcome choice, high risk of bias, and lack of standardized methods that would facilitate the comparison between different clinical trials.81
In two cohorts of moderate-to-severe AD patients, the drug survival of dupilumab (time from initiation to discontinuation of treatment) was compared with other oral immunosuppressive drugs (cyclosporine A and methotrexate) in terms of effectiveness, safety, and patients and doctors’ preferences.82 Dupilumab was found to have a significantly longer drug survival (91% and 88% after 1 and 2 years, respectively) compared to cyclosporine A (37% and 20%) and methotrexate (41% and 33%). Only a small number of dupilumab patients discontinued treatment due to side effects and/or ineffectiveness, while approximately half of the patients discontinued cyclosporin A and methotrexate due to unresponsiveness to treatment.82
In a study including 144 children with AD (age 0–3 years), AD persistence was predicted based on SCORAD at the age of 3 years, low vascular endothelial growth factor serum levels and trigger factors such as stress, change in weather, pollen exposure, infection, and vaccination. Additionally, three distinct endotypes were found: a severe endotype characterized by an early disease onset, high SCORAD, allergic comorbidities, and elevated serum IgE, IL-17, and macrophage inflammatory protein-1b; a benign endotype with late disease onset, lower SCORAD, few allergic comorbidities, and a low concentration of inflammatory serum proteins apart from vascular endothelial growth factor, tissue inhibitor of metalloproteinase-1, and IL-13; and an intermediate endotype.83
Bakker et al. described a biomarker signature named predicted Eczema Area and Severity Index (p-EASI) consisting of a weighted combination of the levels of the serum biomarkers TARC, soluble IL-2-receptor and IL-22. p-EASI can predict disease severity of AD patients treated with dupilumab, topical corticosteroids and cyclosporine A. The use of the p-EASI serum biomarker profile might improve comparisons between clinical trials outcomes and facilitate the assessment of therapy in daily practice.84
Janmohamed et al.85 described an algorithm to be used for AD diagnostic workup in early childhood. A typical morphology and age-specific pattern can be identified in infancy (0–2 years) and early childhood (2–8 years). In early-onset severe AD (<3 months), certain primary immunodeficiency syndromes, genetic disorders with an impaired barrier function, and some metabolic diseases should be considered for differential diagnoses. The diagnostic workup should include assessment and counselling of individual provocation factors and should take into account complications, mainly bacterial or viral infections.85 The proper diagnostic workup helps in choosing the optimal treatment regimen, mostly based on AD severity.86 Daily skincare, avoidance of provocation factors and basic therapy to restore the disturbed skin barrier are always necessary. Mild AD can usually be controlled by reactive therapy with modern topical corticosteroids or topical calcineurin inhibitors, in addition to basic therapy. Moderate-severe AD often requires wet wrap therapy, UV therapy, antiseptics, climate therapy and psychological counselling. In severe AD, systemic corticosteroids might be used, but are not recommended due to unwanted side effects. Long-term H1-antihistamine use may affect sleep quality and is not recommended.86
Recently, Rinaldi et al. demonstrated a new application of electrical impedance spectroscopy (EIS) that is a noninvasive tool for detecting skin barrier integrity in vivo. Specifically, they showed the potential role of EIS for the characterization of the epithelial barrier function.87 The clinical potential of EIS highlighted by these results was further investigated in patients with AD. Using artificial intelligence-based models, EIS could distinguish with good specificity and sensitivity between the skin of persons without AD, and the non-lesional skin of AD patients, and lesional eczema skin from non-lesional skin of patients.87 Also, EIS could assess the skin lesion healing in response to treatment, correlating with the disease score SCORAD and the pruritus score calculated at the same visits, confirming the clinical potential of EIS to objectively assess disease and to follow-up lesions in AD.87
5 BIOMARKERS IN ALLERGIC RHINITIS
With the accumulating evidence and broader implementation of nasal allergen challenge (NAC), local allergic rhinitis (LAR) and non-allergic rhinitis (NAR) are increasingly recognized. Apparently, the combination of clinical history and SPT/sIgE test is not sufficient for the precise diagnosis of AR.88 Therefore, novel biomarkers have been studied including the numbers of several immune cells and various molecules (Figure 3).
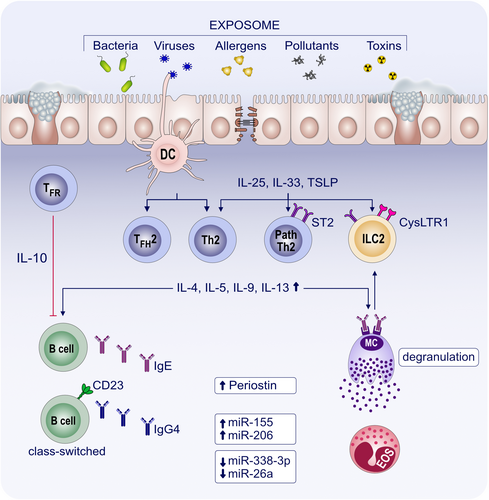
5.1 Immune cells and molecules
Meng et al. comprehensively reviewed novel findings on the role of genetics and environmental factors in the pathogenesis of AR, underlying molecular mechanisms and treatment options. Highlighted biomarkers that are connected to AR are genetic polymorphisms in TSLP (rs1898671) or IL1RL1 (rs3771180), differential epigenetic DNA methylation of, for example, SLFN12, MUC4 or FOXP3, altered miRNA levels of, for example, miR-210 or miR-125, differences in the presence and ratio of cell types such as IL1LR1+ T memory cells and ILC2s, as well as altered concentrations of relevant proteins including IL-17 or IgE. These biomarkers can be indicative of susceptibility, severity or prospective treatment options and responses.89
CD23 expression on B cells is reported as an indicator of disease progression and linked T-B cell bridge. Switched memory B cells from patients with AR expressed higher levels of CD23 which is induced by Tfh2 cells via IL-4. Due to defective IL-10 expression, AR-associated Tfh cells could not sufficiently suppressed IL-4. Allergen-specific immunotherapy (AIT) could significantly reduce the percentage of B cells expressing CD23.90
In addition to T and B cells, ILC2s play a critical role in the pathogenesis of AR by producing a large amount of type 2 cytokines. ILC2s from patients with house dust mite (HDM)-induced AR have a higher expression of cysteinyl leukotrienes receptor compared to healthy subjects91 and were further upregulated in patients with AD.92 CysLT1 expression on the surface of ILC2s may be partially involved in the clinical effectiveness of treatment with the CysLT1 antagonist montelukast in AR and AD.91
While our knowledge of allergen-specific diseases such as AR is growing, there are limited studies on their multimorbidities despite their rather high prevalence. A recent study analyzed the transcriptome of peripheral blood mononuclear cells (PBMCs) of patients with rhinitis, asthma, AD and combinations thereof to investigate the mechanisms of the development of allergies and epigenetic variation and childhood asthma in Puerto Rican patients cohorts. Of note, they identified 8 transcripts (CLC, EMR4P, IL5RA, FRRS1, HRH4, SLC29A1, IL1RL1) that are upregulated specifically when multiple allergic diseases are present.93
5.2 Diagnosis and treatment
Nasal allergen challenge can be used as a safe and reproducible diagnostic tool in both atopic and non-atopic patients. It can diagnose LAR in patients with negative SPT and undetectable serum sIgE. Eguiluz-Gracia et al. enrolled 1165 patients with AR, 369 with non-allergic rhinitis and 361 healthy subjects and performed one or more NACs on all subjects. They showed that changes in volume 2–6 cm (%Vol2–6 cm) determined by acoustic rhinometry alone can rule in and rule out nasal allergen-specific reactivity, and identified a decrease of more than 24.5% as the optimal cutoff point.94
Patients with a positive SPT to seasonal allergens, but with perennial rhinitis symptoms can suffer from either mixed rhinitis (coexistence of AR and NAR) or dual AR (coexistence of AR and LAR). NAC is crucial to differentiate them. In addition to the NAC, BAT could be implemented in the diagnosis of local AR and dual AR.95
Emerging novel biomarkers have advanced precision medicine for the treatment of allergies. Biologics targeting several molecules have been developed, such as benralizumab, dupilumab, mepolizumab, omalizumab and reslizumab. Moreover, AIT can reduce the symptoms in patients with AR and the development of asthma.96 A recent study showed periostin serum levels as a predictive biomarker of response to HDM sublingual immunotherapy (SLIT) in AR patients.97
6 BIOMARKERS IN CHRONIC RHINOSINUSITIS
Chronic rhinosinusitis is a complex disease encompassing two phenotypes, CRSwNP and CRSsNP, based on whether nasal polyps are present or absent. Stratification into additional disease endotypes according to different molecular biomarkers could provide valuable diagnostic information for targeted treatment. These biomarkers include the presence of specific cell populations, proteins, noncoding RNAs, mRNAs, Th2 specific cytokines, cytokines from epithelial cells, epithelial remodeling, and eosinophil and neutrophil counts (Figure 4).98 CRSwNP is often associated with a Th2 response. Zhang et al. analyzed the nasal polyp samples of 21 CRSwNP patients and found higher protein levels of the Th2 cytokines IL-4, IL-5 and IL-13 and the associated inflammatory mediators IgE and ECP compared to controls. They demonstrated that IL-4 and IL-13 stimulated higher MUC5A and MUC5B secretion especially in IL-5(+) CRSwNP samples.99 Another role of IL-13 in CRSwNP is connected to the hippo pathway including the yes-associated protein (YAP). Both factors have been associated with tissue remodeling processes. Yuan et al. provided mechanistic details by showing that IL-13 was able to induce YAP activity, leading to proliferation and differentiation of goblet cells. This process could be reversed by using a YAP inhibitor.100
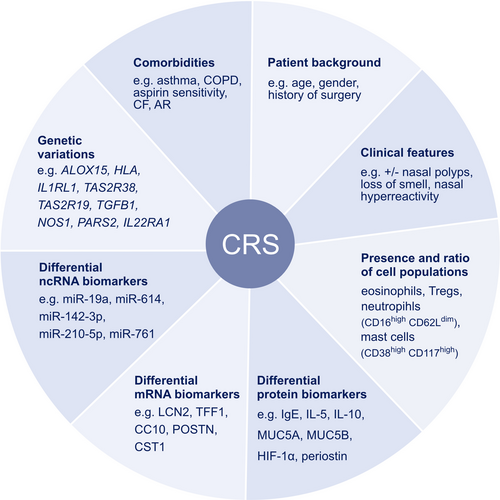
The different CRS endotypes are characterized by different types of cellular and pathological features. Recently, a specific CD38highCD117high mast cell population has been identified in CRSwNP patients that expands during an inflammatory response.101 Mann et al. reported that the percentage of Tregs to CD4+ cells was higher, but less activated, in the nasal tissue of CRSwNP compared to the CRSsNP and control group. Steroid treatment increased the proportion of Tregs in both CRSsNP and CRSwNP, but were more and less activated, respectively. This emphasizes the need to measure activation markers beyond pure assessment of cell types.102 In order to assess the activity of structural cells and immune cells in CRS nasal lavage fluids, microparticle (MP) analysis may be a useful tool. In a recent study, the different subtypes of MPs were characterized based on expressed antigens and their relation to CRS endotypes. Severe CRSwNP was marked by high ITGB6(+)MP levels, indicating basal cell activation.103
Unlike the plethora of research on tissue eosinophilic inflammation for the diagnosis of CRS, there are limited studies on tissue neutrophilia in CRS. Succar et al. provide insights into the inflammatory response in CRS patients in connection to neutrophil counts. The quality of life of CRS patients as measured by the SNOT-22 scores was more affected by the presence of neutrophils than eosinophils.104 Understanding the role of neutrophils might provide additional insights into disease mechanisms of CRS. Arebro et al. characterized the presence of different neutrophil subsets in CRSwNP patients. They showed that CD16high CD62Ldim neutrophils are significantly increased in polyps of CRSwNP patients compared to nasal mucosa both from CRSwNP patients and healthy controls. This cell population has been attributed to be highly activated and is shown to be more efficient in the uptake of S. aureus bioparticles and increased reactive oxygen species production in vitro.105 The pathogenesis of non-eosinophilic CRSwNP was studied by Zhong et al.106 and it was found that HIF-1α mediates the activation of the NLRP3 inflammasome suggesting a role of type 1 polarized macrophages in nasal polyps.
Genetic factors also play a role in CRS, as highlighted in a recent GWAS study in which multiple loci associated with nasal polyps and risk of CRS were identified. Of note, a missense mutation in the ALOX15 locus causing a near total loss of 15-LO enzymatic activity seems to be protective against nasal polyps and CRS, and might be a potential target for treatment. Other genes that are associated with CRS include HLA, IL1RL1, TAS2R38, TAS2R19, TGFB1, NOS1, PARS2 or IL22RA1.107-109 Changes in expression levels of certain transcripts have been associated with CRS, as for example LCN2, TFF1, CC10, CST1 or POSTN and specific miRNAs such as miR-142-3p, miR-210-5p, miR-761, miR-19a or miR-614.110-113 Liao et al.114 identified biomarkers for difficult-to-treat CRS in a Chinese CRS patient cohort by using a multivariate logistic regression model. Characterizing the different disease endotypes of CRS can be valuable to assess different treatment options and tailor them to patients. Hopkins presents decision trees for the diagnosis and treatment of CRS following current clinical recommendations, including stratification according to eosinophil and neutrophil presence and the recommendation for the use of biologics in specific settings.115 Since subtypes of CRSwNP are associated with a Th2 response, Jonstam et al.116 performed a randomized controlled trial on patients with CRSwNP assessing the effects of dupilumab. This resulted in a reduction of the total IgE and eotaxin-3 as well as other type 2 biomarkers. Izuhara et al. reported that periostin, a matricellular protein could act as a biomarker for chronic allergic diseases. Serum periostin levels are significantly elevated in CRS patients and are predictive of treatment response to mepolizumab, lebrikizumab, and postoperative recurrence.117
The association between nasal hyperactivity (NHR) and CRS is not fully understood. Doulaptsi et al. studied the most common triggers in self-reported NHR in CRSwNP and CRSsNP patients, highlighting the differences between the stimuli in the two groups. Humidity changes and emotional stress were found as major triggers in CRSsNP and odors in CRSwNP.118
The COVID-19 pandemic has raised questions for individuals with preexisting respiratory conditions. Multiple studies have analyzed the expression patterns of angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2), the entry receptors of SARS-CoV-2, in different tissues to assess the risk of infection. According to a recent study, ACE2 expression was similar across the sinonasal airway of CRSwNP, CRSsNP patients and healthy controls.119 However, Wang et al. stratified CRS patients according to their inflammatory profile and reported that ACE2 expression upregulated by type 1 cytokine IFN-γ was higher in non-eosinophilic CRSwNP compared to CRSwNP nasal tissue. This emphasized the importance of endotyping CRS patients for personalized treatment.120
7 BIOMARKERS IN FOOD ALLERGY AND ANAPHYLAXIS
In the last decades, food allergies have become a major global health issue. The development of food allergies is a complex process that is regulated predominantly by genetics, immune responses, epithelial function and environmental factors.121 Allergic sensitization to several types of food allergens is mainly caused by an impaired skin barrier or gastrointestinal tract (Figure 5). Multiple innovations have led to the discovery of different biomarkers including basophil and mast cell activation tests, allergen-specific T cells, allergen-specific Igs, microbiome and omics sciences (Figure 6).122

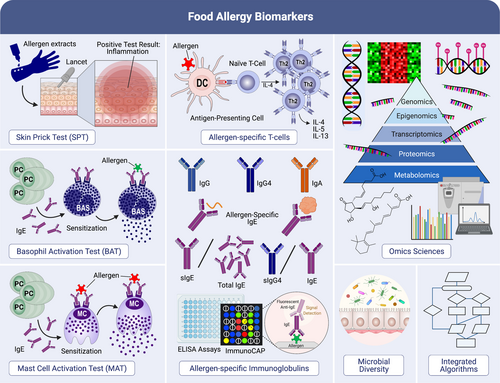
Interestingly, allergen-specific IgE antibodies are mainly produced in human B cells. Satitsuksanoa et al.123, 124 have comprehensively reviewed the fundamental biological functions of B cells in IgE-mediated food allergy. B cells with the capacity of allergen-specific IgE antibody secretion are strongly involved in the development of food allergy and also in maintaining food tolerance. The generation of these allergen-specific IgE antibodies takes place during a combination of B-cell class–switching and affinity maturation. B-cell class–switching controls the expression of different B-cell isotypes including IgG1, IgG2, IgG3 IgG4, IgE, IgA1, or IgA2 heavy chain.123 Allergen-specific IgE antibodies can be sequentially switched through IgG1 isotypes and other isotypes. Jimenez-Saiz et al.125 demonstrated that IgG1+ B cells from an epicutaneous sensitization food-allergic model precede the induction of IgE responses. In addition, B-cell class switching to IgG2 and IgG4 isotypes occurs indirectly following a switch from IgM to the more proximal IgG3 and IgG1 genes rather than directly from IgM to IgG2 or IgG4. Heeringa et al.126 observed increases in allergen-specific IgG2 and IgG4 alongside increased frequencies of IgG2+ and IgG4+ memory B cells, suggesting a class switching of allergen-specific IgG1+ memory B cells upon dual-exposure to allergens. Thus, immune responses against allergens using IgG2 and IgG4 subclasses might ameliorate IgE-mediated allergy. To evaluate the diversity of food allergen-specific IgE, Dang et al.127 measured the level of specific IgE to four egg allergens (Gal d 1, 2, 3 or 5) in the HealthNuts cohort of food challenge-proven egg-allergic, egg-sensitized and egg-tolerant individuals. They found that Gal d 1 sensitization increased the risk of persistent egg allergy by 2.5-fold, and the presence of specific IgEs against all four egg allergens increased the risk of having persistent raw egg allergy four-fold. Therefore, assessing the diversity of IgE-binding to different egg allergen components could be a useful method for prognostic prediction of egg allergy. During the activation of IgE-mediated food allergy, allergen-specific IgE antibodies are cross-linking to FcεRI (FcεRI) on mast cells and peripheral blood basophils, resulting in the secretion of histamine, leukotrienes, and pro-inflammatory cytokines. Monino-Romero et al.128 investigated the function of the soluble isoform of FcεRI in allergen-specific IgE/FcɛRI-mediated activation. Their results indicated that sFcɛRI is released after IgE/FcɛRI complex activation and acts as an endogenous inhibitor of IgE loading to FcɛRI and IgE-mediated activation. Indeed, boosting endogenous soluble FcεRI levels could be a potential treatment for the prevention of IgE-mediated allergic reactions.
In peach allergy, nonspecific lipid transfer proteins (nsLTPs) seem to play a major role during allergen sensitization. These nsLTPs regularly cause severe allergic reactions as they are very stable and unchanged during thermal processing and human digestion. nsLTPs are considered panallergens, a family of related proteins that share highly conserved sequence regions, structure and function, and are present in allergens of various species. Many patients are allergic to nsLTPs not only from a single allergenic source (Pru p 3, peach nsLTP) but to several nsLTPs from different species, which is referred to as LTP syndrome. Recently, Bogas et al.129 demonstrated that most LTP-dependent peach-allergic patients with an early-onset develop LTP syndrome. Hence, there is a current need to find predictive markers of LTP syndrome.
In the Markers Of Nut Allergy Study (MONAS), Duan et al. confirmed the effectiveness of BAT as a diagnostic tool of allergic status against peanut and tree nuts in multisensitized children using %CD63+ basophils in whole blood samples stimulated with nut allergen. Analyzing the BAT results using a mixed-effects Bayesian model, the area under the ROC curve was found to be 0.98 for peanut, 0.97 for cashew, 0.95 for pistachio, 0.92 for hazelnut and 0.97 for walnut. BAT outperformed sIgE testing for peanut or hazelnut and was similar for walnut. In conclusion, BAT was shown to be a reliable diagnostic tool for peanut and nut food allergy in children with multiple sensitizations against several nut species.130
Recent mouse studies have provided mechanistic insights into food allergy, including the modification of epithelial metabolism, induction of regulatory cell subsets and the microbiome. Among traditional (oral) allergen immunotherapy, antigen-independent approaches to treat food allergy are under investigation, namely the use of biologics targeting TSLP, IL-4, IL-13, IL-33 and IgE, and designed ankyrin repeat proteins.131 One additional aspect of relevance is CD300f, an inhibitory receptor expressed on intestinal mast cells. In concert with CD103+ CD11b+ DCs, the interaction of the immunoreceptor CD300f and its ligand ceramide was shown to inhibit the development of OVA-induced food allergy in mice by inhibiting mast cell activation and suppressing Th2 responses.132 Long-acting CD300f agonist drugs, such as ceramide, might therefore be promising for preventing and treating food allergy. The tolerogenic potential of naturally present compounds in the human body should also be considered in molecular mechanisms involved in food allergy. Breast milk has been widely discussed for its protective role against food allergy. There is new evidence that this protective role could be related to the presence of effective concentrations of the short-chain fatty acid butyrate, which modulates several tolerogenic mechanisms involved in food allergy, including modulation of the gut barrier, Treg activation and modulation of Th1/Th2 balance in animal models of food allergy.133
Mondoulet et al. explored the mechanisms of epicutaneous immunotherapy (EPIT) for the treatment of food allergies in murine allergy models, which induces sustained unresponsiveness and prevents further sensitization. They identified EPIT-induced DNA methylation changes with a significant hypermethylation of the Gata3 promoter in Th2 cells and a significant hypomethylation of the FoxP3 promoter in Tregs. This demonstrates that unique and stable changes in the epigenetic signature that downregulate Th2 regulators and upregulate Treg transcription factors are involved in the sustained protective response to EPIT.134
Castan et al. provide a comprehensive overview and critically evaluate the current endpoint parameters used in predictive risk assessment methods in murine food allergy models to test the potential allergenicity of new or modified proteins. They conclude that characterizing a food allergy model using temperature, level of Igs, phenotyping of the cell infiltrate, and cytokine production can provide information on the reaction and the degree of the protein's sensitizing capacity, while the currently available models cannot quantitatively predict the allergic potential.135
The clinical manifestations of food allergies are generally induced by specific IgEs with symptoms ranging from pruritus, wheezing, vomiting, and diarrhea to anaphylaxis. The term “anaphylaxis” refers to a rapid, systemic reaction that can lead to hypotension, respiratory failure, and death. Anaphylactic reactions are not only limited to food-allergic individuals, but can occur as a serious allergic reaction upon contact with some allergens in individuals sensitized to these allergens. Aeroallergens on the other hand do not cause systemic anaphylactic reactions upon inhalation in environmental exposure doses. Local challenges with segmental allergen exposure however can cause anaphylactic reactions locally.136 There are limited studies on the diagnosis, management, and prevention of anaphylaxis. Newer models of adrenaline autoinjectors facilitate the administration of adrenaline. There is a current need to improve public knowledge of using autoinjectors, especially for people at risk of anaphylaxis,137 and for individuals working in day care centers. Blazowski et al. classified clinical phenotypes of real-life food-induced anaphylaxis in a large homogenous children population. Their results reveal two clinical phenotypes of different severity. Importantly, the first phenotype is defined as skin/non-severe respiratory phenotype with mild-to-moderate allergic reactions and no cardiovascular, life-threatening respiratory, and severe central nervous system. The second phenotype is four times less frequent and characterized by a severe respiratory/cardiovascular phenotype, with predominant severe to life-threatening manifestations, including all cardiovascular, severe respiratory, and central nervous system symptoms.138
A recent study highlighted the role of neutrophils in drug-induced anaphylaxis upon administration of neuromuscular-blocking agents (NMBAs) during anesthesia. Neutrophils become activated upon immune complex formation of IgG antibodies against NMBA with administered NMBA This IgG-dependent pathway can aggravate acute hypersensitivity reaction in concert with IgE-dependent immune activation, or can even induce a hypersensitivity reaction without.139
In a study by Vitte et al., serum tryptase, a specific protease secreted after mast cell activation, was assessed as a potential diagnostic biomarker to discriminate between non-anaphylactic and anaphylactic events. In a retrospective study of 102 patients who had experienced a perioperative hypersensitivity reaction, 76 showed clinical diagnostic criteria for drug-induced anaphylaxis based on EAACI guidelines. Among the four medical algorithms tested, the most effective method to discriminate between anaphylactic and non-anaphylactic events was found to follow the 2012 consensus recommendation that acute total tryptase levels are considered significant if greater than [(1.2 × baseline tryptase) + 2] μg/L. Acute mature tryptase measurements were of lower sensitivity and varied according to the severity of the drug-induced anaphylactic reaction.140
As the intensity and development of food-induced anaphylaxis are only partially dependent on the levels of specific IgE, it has been suggested that it is in part regulated by immunomodulators.141 Based on the hypothesis that a defective PGE2 metabolism might be responsible for the development of anaphylaxis, Munoz-Cano et al. investigated the effect of exogenously administered PGE2, the expression profile of PGE2 receptors on basophils, and changes in PGE2 plasma levels. They found that exogenous administration of PGE2 inhibited allergen-induced basophil activation in food-induced anaphylaxis patients and decreased basophil activation through the IgE/FcεRI and N-formylmethionine-leucyl-phenylalanine receptors in both healthy volunteers and anaphylaxis patients. The plasma levels of PGE2 were similar for healthy controls and patients, but food-induced anaphylaxis patients showed a distinct PGE2 receptor pattern on basophils with an elevated expression of pro-inflammatory EP3 over anti-inflammatory (EP2+EP4) receptors.142
Pouessel et al. aimed to identify risk factors for fatal food-related anaphylaxis by analyzing case report data in the allergy vigilance network database.143 They confirmed that peanut and tree nuts are the most frequent food allergens leading to fatal anaphylaxis followed by cow and goat/sheep milk. These findings highlight the challenges of avoidance of trigger foods and the risks involved in oral food challenges. Strategies to avoid fatal anaphylaxis cases need to take into account the young age of patients, additional risk factors, the persistence of mammalian milk allergy, and previous history of food allergies. Prevention and management of food allergies in school settings needs to be improved.144
8 BIOMARKERS OF DRUG HYPERSENSITIVITY
Although there is a high potential of biologicals (mainly mAbs) to treat food allergy, there is a risk of developing hypersensitivity reactions to the biologicals themselves. These hypersensitivity reactions can range from mild to life-threatening symptoms. With an appropriate risk stratification and premedication, rapid desensitization is a novel option to safely re-expose patients to biologicals after a hypersensitivity reaction to maintain first-line therapy. The severity of reaction along with skin testing results is an important guide to decide if desensitization can be performed on the patient.145 During the last 2 years, different assays have been developed to test hypersensitivity to various drugs, improving sensitivity and specificity of in vitro tests as well as new genetic associations related to the risk of severe cutaneous adverse reactions.146 Combining in vitro and in vivo tests is recommended for accurate drug hypersensitivity reaction (DHR) diagnosis. In addition, there is a need to standardize the use of allergy cards to facilitate the communication of patient's drug allergy history to avoid mislabeling of DHRs, a challenge for drug allergic and non-allergic individuals.147 Accurate DHR diagnosis often requires drug provocation tests, which may not be safe for individuals with severe reactions. One of the most challenging diagnoses is drug reaction with eosinophilia and systemic symptoms (DRESS) due to overlapping clinical symptoms at an early stage with drug-induced maculopapular exanthema (MPE). Recently, severe drug-induced MPE after the treatment of severe COVID-19 patients was reported. These patients showed more severe systemic inflammation than DRESS and MPE.148 Interestingly, seven miRNAs were identified as potential biomarkers that correctly identified DRESS or MPE patients.149 The use of omics technologies, such as single-cell RNA sequencing (scRNA-seq), could be a promising new strategy to better understand molecular mechanisms of DHR at the single-cell level. ScRNA-seq on skin and blood samples from a patient with refractory DHR/DRESS were performed and their data pointed to the JAK-STAT signaling pathway as potential target. Subsequent JAK inhibition leads to clinical improvement in this patient.150
Fernandez-Santamaria et al.151 investigated whether the interaction between the checkpoint inhibitor Tim3 and its ligand Gal9 is involved in the drug-induced development of MPEs, which is a non-immediate allergic skin reaction mediated by Th1 cells. The authors found elevated levels of Th1 cells in peripheral blood and in skin biopsies of MPE patients compared to controls, albeit with a lower percentage of Th1 cells expressing the Tim3 receptor. In addition, there was a lower expression of Gal9 in the PBMCs of MPE patients, and of Gal9 and Tim3 in DCs. It is worth noting that the addition of Gal9 to cell cultures suppressed drug-induced proliferation in Th1 and Tim3+Th1 cells, but decreased the proliferation of Tregs in MPE patients. This data suggests that the Tim3-Gal9 axis is involved in the pathogenesis of MPE.151
Drug hypersensitivity reactions can be classified according to the mechanisms involved as immune (allergic), either antibody or T-cell–mediated, and nonimmune (non-allergic) reactions, based on excessive inhibition of specific enzymes or off-target occupation of nonimmune receptors.152 Alternatively, DHRs can be categorized according to the interval between drug administration and the onset of symptoms, that is, immediate (IDHRs) or non-immediate reactions (NIDHRs).152, 153
Drug-specific IgE (sIgE) antibodies are the most validated biomarker for evaluating IDHRs, both in vivo and in vitro. SPT and intradermal test are routinely used and the inclusion of major and minor determinants during skin testing is recommended, as in case of beta-lactams.153 Serum-sIgE immunoassays are limited by the lack of drug-specific IgE’s available and poor sensitivity.154 The quantification of specific activation makers on basophils (mainly CD63 and CD203c) is gaining presence as potential biomarkers, especially when no other immunoassays are available (Figure 7).155 Nevertheless, a survey performed by Torres et al.153 showed that there are considerable deviations from recommendations concerning allergy tests and drug provocation tests in the management of suspected beta-lactam hypersensitivity.
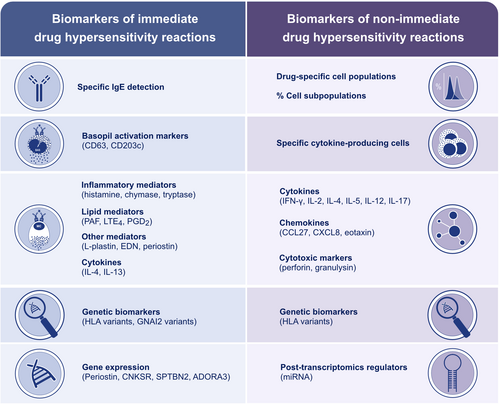
Other important biomarkers for the evaluation of IDHRs include inflammatory mediators (eg, histamine), mast cell proteases (eg, chymase and tryptase), lipid mediators (eg, leukotrienes, prostaglandins, and platelet-activating factor), and cytokines (eg, IL-4 and IL-13) (Figure 7).156 These biomarkers are valuable in evaluating non-allergic reactions in which the detection of Igs is not possible. The assessment of activation markers on basophils is controversial.157
Improvements in genomics have facilitated assessing the relationship between some genetic variants and DHRs. Certain alleles from HLA such as HLA-B*57:01 and HLA-A*31:01 have been associated with severe DHRs induced by abacavir and carbamazepine respectively.152 Similarly, HLA-B*15:02 and HLA-B*58:01 have been associated with the most severe DHRs, such as Stevens-Johnson syndrome and toxic epidermal necrolysis (SJS/TEN).158, 159 In a recent study, Blanca et al.160 reported that GNAI2 variants (rs9883222, rs2298954, and rs2236944) are significant genetic predictors of hypersensitivity reactions to NSAIDs. Recent advances have facilitated the analysis of the complete transcriptome, revealing associations between different gene expression (POSTN, CNKSR, SPTBN2, ADORA3) and NSAIDs hypersensitivity (Figure 7).156
Recently, Sanchez-Lopez et al.161 have reported that 16% of patients with suspected NSAID hypersensitivity could be further diagnosed as suffering from food-dependent NSAID-induced hypersensitivity, with LTP and gliadin being the main culprit food allergens.
Non-immediate reactions are characterized by a wide range of clinical manifestations driven by different mechanisms. The heterogeneity of NIDHR makes it essential to identify biomarkers for accurate diagnosis. In this regard, Torres et al.153 showed that the assessment of drug-specific proliferative lymphocyte response and the detection of cytokine producing cells can represent useful biomarkers to evaluate NIDHRs to betalactams. Similarly, Mori et al.162 reported a lymphocyte transformation test sensitivity of 52% when evaluating 50 children with positive histories of NIDHRs to betalactams. The sensitivity could be significantly improved by measuring the proliferative specific cell subpopulations involved in each reaction.163 The identification of cytokines, chemokines and cytotoxic markers (mainly IFN-λ, IL-5, IL-2, IL-4, perforin and granzyme) released by specific cells also represent important biomarkers for evaluating NIDHRs (Figure 7).152
In a retrospective study, Jakubovic et al. demonstrated the association between IL-6 levels and certain breakthrough reactions that might occur during rapid drug desensitization (RDD) to chemotherapeutic agents and biologicals, and correlated them with clinical manifestations. Elevated levels of IL-6 were found in 76.3% of all of the 38 RDD reactions from 21 patients. Moreover, all the reactions with fever were associated with increased IL-6 (p < 0.0008) and more than half of the reactions including neuromuscular complaints had an increase in IL-6 (p < 0.0042). Additionally, in 19 reactions the mean difference between baseline and reaction IL-6 values was significantly different (1066.8 ± 1257.7, p < 0.0008), showing an increase upon reaction activation. Overall, this data identifies elevated levels of IL-6 as a biomarker of immediate drug hypersensitivity with a characteristic phenotype and suggests the potential of anti-IL-6 therapies to improve the safety of drug administration.164
9 BIOMARKERS IN ALLERGEN IMMUNOTHERAPY
It is well-established that AIT induces allergen-specific Treg and Breg cell responses and increases in their numbers in vivo. Furthermore, an increase in serum allergen-specific IgG4 levels, decreased tissue numbers of mast cells and eosinophils in the affected tissues and basophil as well as mast cell desensitization have been consistently reported.165 Different types of AIT have shown similar effects in this context. In addition, decreased ILC2 numbers and increased ILCreg numbers accompany these findings.166, 167 Various biomarkers are used to measure the immune response and efficacy of AIT (Figure 8). In terms of immune cells as biomarkers in AIT, inducible IL-10+ type 1 Treg cells show an early response in low-dose AIT and are crucial to the induction and maintenance of peripheral allergen tolerance enhanced by AIT,168, 169 while the percentage of dysfunctional allergen-specific ILT3+ Treg cells and allergen-specific Th2 cells were significantly reduced after AIT treatment.169 Allergen-specific CD45RBlowCD27−CRTH2+CD161+CD49d+CD4+ T cells (Th2A) were found to be more frequent in allergic patients than in control subjects and their decrease indicated a positive outcome of AIT.170
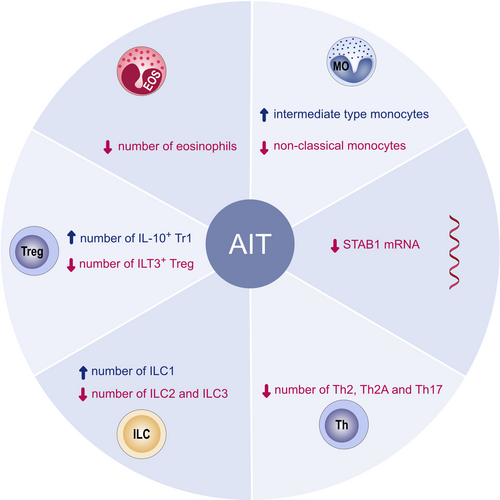
Yao et al.3 provided a comprehensive review on the role of follicular helper and regulatory cells in allergic diseases and the latest research on the mechanisms by which AIT is able to balance their functions. Schmid et al. found that basophil activity was reduced 447-fold after the first year of subcutaneous immunotherapy (SCIT) treatment, remained 100-fold lower than initial values in the treatment period of 3 years, and was 10-fold lower than initial values in the first year of follow-up. From these findings, they proposed that decreased basophil sensitivity predicts long-term improvement in the clinical outcome by SCIT.171
In a double-blind placebo-controlled clinical HDM AIT study, Claire et al.172 identified 14 potential gene markers involved in allergic responses and proposed IL-10 mRNA abundance as a predictive biomarker of an effective clinical response to AIT. Serum proteins such as IgE, allergen-specific IgG1, allergen-specific IgG4, cytokines, chemokines and serum inhibitory activity for IgE such as IgE-FAB or IgE-BF are also convenient biomarkers to monitor the clinical improvement of AIT.173
The first years of life are critical in the development of allergic diseases. Zhang et al.174 studied the changes in DNA methylation in adolescent individuals pre- and post-sensitization, and identified 35 CpG methylation sites, one of which strongly correlated with increased levels of IL5RA in peripheral blood. Moreover, Schmitt et al.175 performed a longitudinal study in 4111 asthmatic patients showing that AIT treatment reduced disease progression, particularly in younger individuals. Similarly, Luce et al.176 studied the effects of omalizumab and multi-oral immunotherapy in young individuals suffering from food allergy. Multi-oral immunotherapy was found to increase the levels of FcγRIIIa, and patients older than 10 years of age exhibited a concomitant reduction of Th2a and Th17 cells and STAB1 mRNA expression.
Allergen-specific immunotherapy has been successfully demonstrated to induce immune tolerance against the culprit allergen in adults. Barker-Tejeda et al.177 studied the use of novel omic biomarkers for sublingual immunotherapy (SLIT) treatment in grass pollen allergy. After 2 years of treatment, the data indicates that SLIT had only resulted in desensitization of effector cells. This has been suggested as a possible reason for the loss of the clinical effect after discontinuation of a 2-year treatment. In another cohort, Varona et al.178 monitored patients undergoing a 3-year SLIT treatment during a 5-year period. Successful treatment was found to be associated with the induction of a regulatory response marked by higher numbers of Tregs, lower eosinophils and IgE during pollen season. Immunological changes resulting from AIT are not limited to the adaptive response, Eljaszewicz et al.179 showed that grass and birch AIT was able to restore the composition and numbers of ILCs, DCs and monocytes to the levels of non-allergic individuals. A new AIT formulation using peptides provided similar immune responses in cat-sensitized AR individuals.180 Total nasal symptom scores (TNSS) were reduced in all treated individuals at all the timepoints measured. Furthermore, there was a significant reduction in the neutrophil to lymphocyte ratio 1–2-hour post-challenge and the number of lymphocytes correlated negatively with TNSS.
In a retrospective study, Wahn et al.181 analyzed the German prescription database LRx to compare AIT patients (n = 9001 with six different variations of allergen formulations) with non-AIT patients (n = 45,005) as a proxy for clinical status and disease progression of pollen-associated AR and asthma. Data contained longitudinal information from discontinuation of AIT to up to 6 years of follow-up. The AIT and non-AIT patients were matched to have comparable distributions for statistical analysis. Data were integrated into a general linear model with the ratio of annual subscriptions throughout follow-up versus the year prior to receiving the first dosage of AIT medication as the outcome variable. The analyses revealed a significant (p < 0.001) decrease in AR medication usage in the AIT group compared to the non-AIT group. Moreover, the AIT group showed a reduction in asthma medication use in patients with present asthma (p < 0.001). Overall, 49.1% of the AIT group were asthma medication-free in comparison to 35.1% of the non-AIT group. In addition, AIT also significantly reduced the risk of new-onset asthma (p < 0.001).
Adjuvants are essential components of the AIT vaccine as they stimulate the innate immune system and provide a stable reservoir of antigen at the site of injection. The effect of an adjuvant can be considered as an important biomarker of the efficacy of AIT, as some adjuvants such as allergoids conjugated to mannan are capable of increasing allergen uptake and promote the induction of Th1 and Treg cells.182, 183 Sialic acid-modified antigens promote immune tolerance by instructing DCs to induce de novo Tregs and inhibit the generation of inflammatory T cells.184 Sialylation levels of allergen-derived peptides were also used as a biomarker of peptide SCIT efficacy.185
Certain medical scores or classifications can be used as markers of AIT outcome. In a meta-analysis of randomized controlled trials, Yanran et al.186 adopted nasal symptom scores, medication scores and prevalence of adverse effects as indicators to assess the efficacy and safety of HDM SCIT for AR. The Global Initiative for Asthma classification187 was used as marker of asthma severity and it was found that in patients who receive AIT the likelihood for the progression of asthma severity is reduced.188 Furthermore, the cost-effectiveness of SCIT plus ICS versus ICS alone for the treatment of allergic asthma was compared.189 Biomarkers related to e-health or mobile health and a clinical decision support system of AR and asthma may also be useful as indicators in AIT and help patients' disease management.190, 191 Within the same context, Pfaar et al.192 proposed a clinical algorithm for AIT decision-making and optimal clinical practice. This algorithm follows the latest recommendations from the EAACI guidelines.
10 BIOMARKERS IN COVID-19 PATIENTS COMBINED WITH ALLERGIC DISEASES
The global spread of COVID-19 has raised questions on the diagnosis and management of allergic diseases during these times. The airway epithelium is especially abundant in ACE2 receptors and serves as a gate for SARS-CoV-2 infection.193, 194 Respiratory diseases are broadly discussed for their potential involvement in COVID-19 severity and outcomes. To date, allergic diseases, such as allergic rhinitis, allergic asthma, atopic dermatitis, urticaria, or drug and food allergy, are not considered a COVID-19 risk factor and allergic patients have been reported to have a relatively mild disease.27, 193, 195 Several studies have demonstrated that underlying allergy status in COVID-19 patients led to less severe disease and a lower degree of damage in the lungs.196 Interestingly, allergic inflammatory airway diseases negatively correlated with ACE2 expression.193, 197, 198 Milder course of COVID-19 in allergic individuals was also accompanied by increased frequencies of T cells,199 and eosinophils,200 when compared to non-allergic COVID-19 age-matched control groups. Decreased ACE2 receptor expression and enhanced numbers of lymphocytes and eosinophils in the allergic patients might contribute to the ameliorated infection, more efficient virus clearance, resulting in a less severe outcome. Importantly, lymphopenia and eosinopenia are considered markers of critical COVID-19.195, 201
Allergic asthma is the most common inflammatory phenotype of the disease and is characterized by excessive type 2 immune responses to allergens.202 Similarly to other allergic diseases, allergic asthma is not considered as a risk factor for COVID-19.195, 203 Asthma status was not related to delayed SARS-CoV-2 viral clearance in COVID-19 patients.203 Chang et al.204 investigated a group of 178 asthmatic patients in Beijing. At the time of publication, the majority of asthma patients (89.3%) self-reported well-controlled disease.204 Interestingly, during the same period, more patients experienced asthma exacerbation (25.6% vs. 15.5% previously reported).204 Excessive asthma aggravation might be partially attributed to pollen and other seasonal respiratory viruses, and a reduction of medication intake in 45.8% of patients.204 The latter was implemented as part of COVID-19 measures to ensure the availability of medication during the pandemic. Despite higher exacerbation occurrence, the frequency of visits to the hospital decreased to prevent possible SARS-CoV-2 infection.204 Notably, an EAACI Asthma Section Survey demonstrated that COVID-19 pandemic lockdown decreased quality of healthcare provided to patients, resulting in worsened asthma control.205 In contrast to type 2 asthma, Zhu et al.206 demonstrated that non-allergic asthma was related to an increased risk of severe COVID-19.
Several recommendations and guidelines have been published regarding the management of allergic diseases during the COVID-19 pandemic (Table 2).27, 207 In a EAACI position paper, Klimek et al.27 recommend continuing personalized intranasal corticosteroid treatment in CRS patients. A consensus statement written by the ARIA-EAACI Task Force provides recommendations for asthma and COVID-19 management.207 Continuation of prescribed therapy, proper control of asthma and other allergic diseases is necessary, as a severe and often uncontrolled disease may contribute to the higher frequency of life-threatening exacerbations.27, 198 Biological therapies are used to treat severe cases of type 2 diseases. Biological treatment in non-infected patients during the COVID-19 pandemic should be continued. Once SARS-CoV-2 infection is confirmed, biologicals should be stopped and proper baseline disease control should be assured. After patient recovery, biological treatment should be continued. Any activities leading to disease deterioration should be avoided to minimize doctor/hospital visits and risk of SARS-CoV-2 infection.27, 207 Respiratory symptoms developed upon allergen exposures may be a source of distress and concern during the COVID-19 pandemic, particularly during pollen seasons (Figure 9). The source of observed manifestations (COVID-19 vs. allergy) should always be consulted with a physician (preferably teleconsultation) and individually assessed with regards to the patient's status and comorbidities. It should be noted that in pollen allergic patients, nasal symptoms can also develop during COVID-19 at the times of high-dose pollen exposure, which was not observed in COVID-19 patients during the first months of the pandemic. Extensive screening for SARS-CoV-2 is recommended to help distinguish COVID-19 symptoms from exacerbations of a different origin.207 Additionally, severe allergies are not a contraindication for COVID-19 vaccination for patients without a prior history of allergies to the vaccine components.208, 209
| General recommendations | |||
| Termination of biological treatment is not generally necessary | |||
| Based on an individual risk-benefit analysis, especially in severe allergic patients, biologicals should be continued as scheduled | |||
| Prolongation of the injection interval should be considered to limit the physician-patient contacts to a minimum | |||
| With adequate patient training, self-administration of biologicals should be considered | |||
| Recommendations for patients | |||
| PCR negative asymptomatic individuals | During quarantine period | Mild-to-moderate COVID-19 cases or infection suspicion | Severe COVID-19 cases |
|
Biological therapy can be started for approved indications or can be continued during the current COVID-19 pandemic |
Telemedical support should be considered Continuing the basic therapy with inhaled bronchodilators topical steroids, antihistamines, etc recommended |
Biologicals can be continued regarding respective indications If an individual risk-benefit analysis supports the continuation If the patient consent was given after being informed about the limited availability of data |
Consideration should be done for Prolongation of the injection interval or treatment interruption regarding respective indications Possible requirement of systemic glucocorticosteroids |
- Abbreviations: COVID-19, coronavirus disease 2019; PCR, polymerase chain reaction.
In summary, allergic diseases and asthma do not aggravate the risk of severe/critical COVID-19 outcomes in the absence of underlying lung disease or any other immunosuppressive or metabolic comorbidities.210 Adherence to recommended social distancing and the use of personal protective equipment, such as face masks, can lower the incidence of exacerbations by reducing aeroallergen exposure.200 Patients with any underlying allergic disease should continue with standard treatment and be reassured of its safety and efficacy.27 Detection of SARS-CoV-2 infection in allergic patients might be challenging due to overlap of allergy and COVID-19 symptoms and reluctance for doctor consultations. Screening for SARS-CoV-2 is imperative to minimize viral transmission and to differentiate between COVID-19 symptoms and allergies.
11 CONCLUSIONS
The assessment and management of allergic diseases is challenging since the pathological molecular mechanisms are complex and heterogeneous in their clinical presentation. Improvement of disease management requires a personalized treatment regimen, which requires well-defined and reliable biomarkers to unravel the phenotypes and endotypes of allergic diseases. There is a current need to identify prognostic biomarkers to guide the selection of appropriate therapies and to monitor treatment efficacy. Several biomarkers have been studied in allergic diseases, but only a few are readily available for clinical use. Newly identified biomarkers based on omics and novel imaging techniques must be validated and standardized before they can be routinely applied in clinical practice. The development of robust biomarkers will facilitate precision medicine-based treatment approaches for allergic diseases.
ACKNOWLEDGMENT
We would like to thank Dr Anna Globinska for assistance in generating the figures. Open Access Funding provided by Universitat Zurich.
CONFLICT OF INTEREST
Ismail Ogulur, Yagiz Pat, Elena Barletta, Lacin Cevhertas, Ruben Fernandez-Santamaria, Mengting Huang, Manal Bel Imam, Jana Koch, Siyuan Ma, Debbie J. Maurer, Yaqi Peng, Urszula Radzikowska, Arturo O. Rinaldi, Juan Rodriguez-Coira, Pattraporn Satitsuksanoa, Stephan R. Schneider, Alexandra Wallimann, Damir Zhakparov, Reihane Ziadlou, Marie-Charlotte Brüggen, Katja Baerenfaller, Luo Zhang, Mubeccel Akdis declare no conflict of interest. Yasutaka Mitamura and Willem van de Veen report grants from Novartis. Milena Sokolowska reports grants from Swiss National Science Foundation, GSK and Novartis. Cezmi Akdis reports grants from Allergopharma, Idorsia, Swiss National Science Foundation, Christine Kühne-Center for Allergy Research and Education, European Commission's Horison's 2020 Framework Programme, Cure, Novartis Research Institutes, Basel, Astra Zeneca, Switzerland, Scibase, Stockholm, advisory role for Sanofi/Regeneron, Glaxo Smith-Kline, Novartis.




