Natural influenza infection produces a greater diversity of humoral responses than vaccination in immunosuppressed transplant recipients
Cedric Hirzel and Andrzej Chruscinski share first authorship.
Atul Humar and Deepali Kumar share senior authorship.
Authors of “Influenza in Transplant Study Group” are listed in Appendix A1.
Abstract
The humoral immune response to influenza virus infection is complex and may be different compared to the antibody response elicited by vaccination. We analyzed the breadth of IgG and IgA responses in solid organ transplant (SOT) recipients to a diverse collection of 86 influenza antigens elicited by natural influenza A virus (IAV) infection or by vaccination. Antibody levels were quantified using a custom antigen microarray. A total of 120 patients were included: 80 IAV infected (40 A/H1N1 and 40 A/H3N2) and 40 vaccinated. Based on hierarchical clustering analysis, infection with either H1N1 or H3N2 virus showed a more diverse antibody response compared to vaccination. Similarly, H1N1-infected individuals showed a significant IgG response to 27.9% of array antigens and H3N2-infected patients to 43.0% of antigens, whereas vaccination elicited a less broad immune response (7.0% of antigens). Immune responses were not exclusively targeting influenza hemagglutinin (HA) proteins but were also directed against conserved influenza antigens. Serum IgA responses followed a similar profile. This study provides novel data on the breadth of antibody responses to influenza. We also found that the diversity of response is greater in influenza-infected rather than vaccinated patients, providing a potential mechanistic rationale for suboptimal vaccine efficacy in this population.
Abbreviations
-
- CV
-
- coefficient of variation
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- FBS
-
- fetal bovine serum
-
- HA1
-
- hemagglutinin A1 subunit (hemagglutinin globular head protein)
-
- HA2
-
- hemagglutinin A2 subunit (hemagglutinin stem protein)
-
- HI
-
- hemagglutinin-inhibiting
-
- HIA
-
- hemagglutinin inhibition assay
-
- IAV
-
- influenza A virus
-
- IQR
-
- interquartile range
-
- M1
-
- matrix 1 protein
-
- MFI-B
-
- median fluorescent intensity minus local background
-
- NA
-
- neuraminidase
-
- NP
-
- nucleoprotein
-
- NS1
-
- non-structural protein 1
-
- NS2
-
- non-structural protein 2
-
- PA
-
- polymerase acidic protein
-
- PB1
-
- polymerase basic protein 1
-
- PB2
-
- polymerase basic protein 2
-
- PBS
-
- phosphate-buffered saline
-
- RNA
-
- ribonucleic acid
-
- SAM
-
- significance analysis of microarray
-
- SD
-
- standard deviation
-
- SOT
-
- solid organ transplant
1 INTRODUCTION
Influenza virus infection causes significant morbidity and occasional mortality in the solid organ transplant (SOT) population.1, 2 A key aspect of the immune response includes the development of antibodies directed against specific proteins within the influenza virus.3-7 Because of the immunosuppression required to avoid graft rejection, SOT recipients tend to have reduced antibody responses to both natural influenza infection and to vaccination.8, 9 In the vast majority of studies, vaccine response is measured primarily against the hemagglutinin (HA) protein of influenza. Based on this, transplant patients generally show variable but suboptimal vaccine immunogenicity and they may still commonly acquire influenza despite vaccination. Less data are available on the humoral immune response to natural infection in transplantation but these also show a limited seroconversion when IgG antibodies are measured using a hemagglutination inhibition assay.8
Influenza virus is a member of the Orthomyxoviridae family. The influenza A (IAV) viral genome consists of eight negative-sense single-stranded RNA fragments, which code for different viral proteins, including HA, neuraminidase (NA), matrix proteins (M1 and M2), polymerase protein subunits (PB1, PB2, and PA), nucleoprotein (NP), and non-structural proteins (NS1 and NS2).10 The two major transmembrane glycoproteins of the virus, HA and NA, show continuously shifting amino acid sequences and are therefore antigenically highly variable.11 Other parts of human pathogenic IAVs, such as the external domain of the M2 protein (~90% conserved among IAVs), NP (~90% conserved), M1 (>90% conserved), NS1 (>90% conserved), and NS2 (~90% conserved), are much less variable.11-15 Traditionally humoral immune responses to infection and vaccination against HAs have been studied extensively because these antibodies (hemagglutinin-inhibiting [HI] antibodies) correlate with protection from infection.16 However, influenza vaccination or infection may also elicit antibody responses to influenza proteins other than strain-specific HA proteins.11 For instance, mouse studies show that the presence of anti-NA antibodies was correlated with milder disease, and anti-M2 antibodies can reduce influenza viral loads and are associated with better survival after viral challenge.11, 16
Antigen microarrays can be a useful tool for serologic profiling of humoral immune responses to a range of influenza antigens.17-22 The array technique (as opposed to ELISA) allows for multiplex testing against many different influenza antigens using only small volumes of serum. Two-color detection protocols can also be used to simultaneously measure two antibody classes (e.g., IgG and IgA) on the same slide surface.
There are limited data on the diversity of influenza-specific antibody responses in SOT recipients. We hypothesized that natural infection would likely exhibit a much more robust and diverse antibody response as compared to influenza vaccination. This would partly help explain the suboptimal vaccine efficacy observed in transplant patients. It would also be important to understand the development of antibodies against conserved viral epitopes, and heterologous strain proteins, to help delineate how both vaccination and infection may provide broader protection against other influenza strains. Therefore, the aim of our study was to investigate the diversity of anti-influenza antibody responses to natural infection and vaccination in SOT recipients and to compare differences in humoral immune responses after vaccination and natural infection.
2 METHODS
2.1 Patients and sample collection
Two groups of adult (aged ≥18 years) SOT recipients were included in this study: (1) patients with IAV infection and (2) patients who were vaccinated with one dose of inactivated trivalent influenza vaccine.
Serum samples from IAV-infected SOT recipients (40 patients with influenza A/H1N1 infection and 40 patients with influenza A/H3N2 infection) were obtained from a prospective multicenter cohort study conducted at 20 different centers in the United States, Canada, and Spain during the 2010–2015 influenza seasons.1 Samples were chosen if they had an adequate amount of serum at two time points. All patients had laboratory-confirmed (by center-specific nucleic acid amplification testing) diagnosis of influenza A infection. Serum samples were drawn at enrollment and 4 weeks after.
Serum samples from vaccinated patients (n = 40) were obtained from a double-blind randomized controlled influenza vaccine trial.23 Similar to the infected patients, samples with enough serum to perform the study were chosen. Patients were vaccinated with the 2016–2017 trivalent influenza vaccine Fluviral® (GSK, Canada), which is a split virus vaccine. The vaccine contained antigens (15 µg per antigen) of the following three viruses: A/California/7/2009 (H1N1)pdm09-like virus, A/Hong Kong/4801/2014 (H3N2)-like virus, and B/Brisbane/60/2008-like virus. Sera were drawn immediately before and 4 weeks after vaccination.
2.2 Antigen library
A diverse collection of influenza antigens (n = 86), including those from various subtypes (influenza A/H1N1, A/H3N2, A/H5N1, and influenza B) and geographical locations were used. The microarray consisted of 26 A/H1N1 antigens (14 targeting HA, four NA, and eight targeting conserved antigens), 35 A/H3N2 antigens (27 targeting HA, two NA, two whole viral particles, and three targeting conserved antigens), six A/H5N1 antigens (three targeting HA, one NA, and two conserved antigens), and 19 influenza B antigens (14 targeting HA, one NA, three whole viral particles, and two conserved antigens). A complete list of all antigens arrayed is provided in the Supporting Information (Table S1). Antigens were diluted to 0.2 mg/ml in PBS and stored in aliquots at −80°C until the day of microarray printing.
2.3 Antigen Microarrays
The influenza antigen microarray was generated using previously published protocols for generation of antigen microarrays to screen for autoantibodies in heart failure and transplantation.24-26 Antigens including whole virus lysates, proteins, and peptides were spotted in duplicate onto two-pad nitrocellulose-coated slides (Oncocyte® SuperNOVA, Grace Bio-Labs, Bend, OR) using a VersArray Chipwriter Pro microarrayer (Virtek Vision, Waterloo, ON, Canada). Slides were arrayed at room temperature at a relative humidity of 55%. Solid pins (Arrayit, Sunnyvale, CA) were used to generate features of approximately 500 µm in diameter. Dried slides were placed in FAST frames (Maine Manufacturing, Sanford, ME) and blocked overnight at 4°C (blocking buffer: PBS, 5% FBS, 0.1% Tween). The next day, arrays were incubated with patient serum (diluted 1:100 in blocking buffer) for 1 hour at 4°C. Baseline and convalescent (or postvaccination) serum from same patient were tested on separate pads from the same slide. Thereafter, slides were washed extensively (PBS with 0.1% Tween) and probed and incubated for 45 minutes at 4°C with a mixture of secondary antibodies consisting of Cy3-labeled goat anti-human IgG (Jackson ImmunoResearch, West Grove, PA) at a dilution of 1:2000 and Alexa Fluor 647-labeled goat anti-human IgA (Jackson ImmunoResearch, West Grove, PA) at a dilution of 1:1000. After washing, slides were dried by centrifugation (220 x g for 5 minutes).
Fluorescent intensities of features were quantified using an Axon 4200A microarray scanner (Molecular Devices, Sunnyvale, CA) and GenePix 6.1. software (Molecular Devices, San Jose, CA). Median fluorescent intensity minus local background (MFI-B) was determined (at 532 nm for Cy3 and 635 nm for Alexa Fluor 647). The single averaged MFI-B for each antigen was calculated from the features arrayed in duplicate (for illustration of the antigen microarray, see Figure S1).
2.4 Quality control
The interslide variation (variability of signal within arrays tested the same day) was determined by measuring the variation coefficient of seven slides (14 nitrocellulose pads), which were probed with the same serum sample and processed the same day. The mean CV (±1 SD) was 7.6% (±3.9) for IgG signals and 15.2% (±10.7) for IgA signals. Linearity of the assay was assessed by plotting log2-transformed signals for the different antigens from serially diluted sera (example shown in Figures S2 and S3).
To reduce any impact of signal variability in the analysis, we decided to compare fold changes in MFI-B (MFI-B after 4 weeks/MFI-B at baseline) instead of focusing on absolute MFI-B values. This was possible because of the linearity of the assay and the fact that baseline and 4-week serum samples of the same patient were probed on the same slide. Seroconversion was defined as a ≥ 4-fold increase in MFI-B.
2.5 Hemagglutination inhibition assay (HIA)
For validation of array against a standardized HIA, HI antibody titers for H1N1 A/California/07/2009 were determined for all 120 samples used in the study. HIAs were performed at a World Health Organization (WHO) national influenza reference laboratory (Public Health England) using the methods described previously.27 Titers were determined in duplicate by doubling dilutions of serum using an initial dilution of 1:10. Antibody concentrations below the lower limit of detection (<1:10) were assigned a titer of 5 for the purposes of analysis. Seroconversion was defined as a ≥ 4-fold increase in HI antibody titer.
2.6 Statistical analysis
Demographic characteristics between IAV-infected SOT recipients and vaccinated patients were compared using descriptive statistics (Chi-squared test or Fisher's exact test for categorical variables; Mann-Whitney U test for continuous variables).
Differences in antigen reactivity between baseline and 4-week serum samples were identified as follows: we explored if log2-transformed MFI-B fold changes were different from zero by using Significance Analysis of Microarrays (SAM) with a false discovery rate of <1% (q-value <0.01).28 Only antigen reactivities with a q-value of <0.01 that changed at least 2-fold were considered to be significant.
In order to compare antibody responses of A/H1N1-infected versus vaccinated patients against H1N1 antigens, log2-transformed MFI-B fold changes of A/H1N1 antigen reactivities were compared using SAM with a false discovery rate of 1% (q-value <0.01). The same approach was applied for A/H3N2-infected versus vaccinated patients. Following SAM analysis, hierarchical clustering (Manhattan clustering) of significantly different antigen reactivities was performed using Cluster 3.0 and heatmaps were generated with Treeview 1.60.29 SAM analysis was performed using an Excel add-in.28 Descriptive statistics were done using Stata software version 12.0 (College Station, TX).
3 RESULTS
3.1 Cohort description
A total of 120 patients were analyzed. Baseline demographics are shown in Table 1. Almost half of the cohort were kidney transplant recipients (48.3%). Kidney transplant recipients were more represented in the IAV-infected population (57.5%) compared to the vaccine group (30.0%); p = .006. The proportion of the other organ types between the two groups was not statistically different, although there was a trend to a higher proportion of heart or lung transplant patients in the vaccine group. However, the overall immunosuppression regimen was very similar across groups. Most patients were on calcineurin inhibitor-based immunosuppressive regimens (95.0%); six patients had combined calcineurin inhibitor/sirolimus immunosuppression. All IAV-infected transplant recipients were treated with oseltamivir.
| Characteristic | All (n = 120) | Influenza A infected (n = 80) | Influenza vaccinated (n = 40) | p value |
|---|---|---|---|---|
| Age (years), median (IQR) | 55.5 (45–63) | 55 (44.5–65) | 57 (49.5–62.5) | .911 |
| Male sex | 82 (68.3%) | 51 (63.8%) | 31 (77.5%) | .127 |
| Time from transplantation to infection/vaccination, years (IQR) | 3.0 (0.8–8.1) | 2.8 (0.4–9.6) | 3.3 (1.0–7.4) | .515 |
| Within 1 year of transplantation | 39 (32.5%) | 28 (35.0%) | 11 (27.5%) | .408 |
| Type of transplant | ||||
| Kidney | 58 (48.3%) | 46 (57.5%) | 12 (30.0%) | .006 |
| Liver | 25 (20.8%) | 16 (20.0%) | 9 (22.5%) | .813 |
| Lung | 17 (14.2%) | 9 (11.3%) | 8 (20.0%) | .266 |
| Heart | 10 (8.3%) | 4 (5.0%) | 6 (15.0%) | .082 |
| Intestine | 1 (0.8%) | 1 (1.3%) | 0 (0.0%) | 1.000 |
| Combined | 9 (7.5%) | 4 (5.0%) | 5 (12.5%) | .185 |
| Immunosuppression | ||||
| Prednisone | 93 (77.5%) | 64 (80.0%) | 29 (72.5%) | .354 |
| Prednisone dose, mg/day (IQR) | 5 (5–10) | 5 (5–10) | 5 (3.8–8.8) | .532 |
| Tacrolimus | 77 (64.2%) | 49 (61.3%) | 28 (70.0%) | .346 |
| Cyclosporine | 37 (30.8%) | 26 (32.5%) | 11 (27.5%) | .576 |
| Mycophenolate mofetil/mycophenolate sodium | 88 (73.3%) | 60 (75.0%) | 32 (80.0%) | .559 |
| Azathioprine | 8 (6.7%) | 3 (3.8%) | 5 (12.5%) | .070 |
| Sirolimus | 11 (9.2%) | 10 (12.5%) | 1 (2.5%) | .074 |
| Antithymocyte globulin within 6 months prior | 6 (5.1%) | 5 (6.4%) | 1 (2.5%) | .662 |
| Rejection within 6 months prior | 4 (3.3%) | 4 (5.0%) | 0 (0.0%) | .300 |
- Abbreviations: IQR, interquartile range; mg, milligram.
3.2 Antibody response to influenza infection and vaccination
3.2.1 Antibody response in influenza A/H1N1-infected transplant patients
Of the 86 antigens coated on the microarray, 26 were influenza A/H1N1 antigens (Table S1), including 14 complete HA proteins and eight peptides corresponding to the globular head of HA (HA1). When averaged across all 40 patients, influenza A/H1N1 infection resulted in a significant IgG immune response (mean of >2-fold change in fluorescence) against several HA antigens from different strains of H1N1 (Figure 1; Table S2) as well as other antigens. A total of 27.9% (24/86) of the antigens included in the microarray (Figure 1A) showed a mean 2-fold or higher change in reactivity across the 40 patients. Of these, the immune response was mainly directed against influenza A/H1N1 antigens (75.0%; 18/24). However, heterosubtypic immune responses targeting six H3N2 or H5N1 antigens were also identified. Of note, five of these six antigens were either matrix 1 (M1), NPs, or a HA stem protein (HA2) (Table S2). All these proteins are considered well-conserved influenza A proteins.11
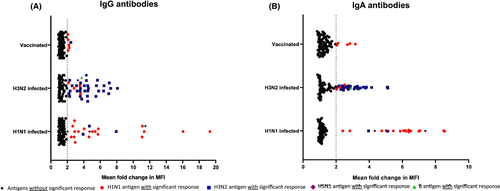
Significant IgA responses were identified against 23.3% (20/86) of the antigens (Figure 1B). Again, the immune response was mostly directed against influenza A/H1N1 antigens (80.0%; 16/20) (Table S2). The majority (75.0%; 3/4) of the significant IgA antibody responses against non-A/H1N1 antigens were directed against conserved influenza A antigens (two M1 proteins and a HA2 protein). All antigens with a significant IgA response were also significant for IgG antibody response. However, 16.7% (4/24) of antigens for which we found significant IgG immune responses did not show IgA antibody responses (Table S2).
3.2.2 Antibody response in influenza A/H3N2-infected transplant patients
The antigen microarray included 35 different influenza A/H3N2 antigens (including 11 complete HA proteins, and 15 HA1 peptides; Table S1). When averaged across all 40 patients with H3N2 infection, a significant IgG response was detected against 43.0% (37/86) of the antigens included in the microarray (Figure 1A). The antibody response was mainly targeting A/H3N2 antigens (83.8%; 33/37) (Table S3). Heterosubtypic responses to six non-A/H3N2 antigens were present, and mainly targeted conserved IAV antigens including three M1 proteins, one NP protein, and one NS1 protein (Table S3).
IgA antibody responses were present for 37.2% (32/86) of the antigens (Figure 1B). The IgA response was primarily detectable against A/H3N2 antigens (81.3%, 26/32) (Table S3). Most of the antigens against which we detected significant IgA antibody responses also showed significant IgG responses (90.6%; 29/32). However, for 21.6% (8/37) of antigens with significant IgG immune responses, there was no significant IgA immune response (Table S3).
3.2.3 Antibody response to trivalent influenza vaccine
The diversity of antibody responses in vaccinated transplant recipients was poor. When averaged across 40 patients, significant IgG antibody responses were observed to only 7.0% (6/86) of the antigens (Figure 1A). The vaccine contains H1N1, H3N2, and B viruses (and was a split virus vaccine preparation). However, responses were only observed against IAV and predominantly H1N1 antigens (5/6 83.3%). Three of the responses were against the H1N1 HA1 protein (the globular head) while three were against the M1 protein (two H1N1 and one H3N2) (Table S4).
Significant IgA responses were detected for 5.8% (5/86) of the antigens (Figure 1B). These antigens were either complete HA proteins or the HA1 globular head protein (Table S4). Three of five antigens (60.0%) with significant IgA responses also had significant IgG antibody responses. For half (3/6; 50.0%) of antigens with IgG immune responses, there was no significant IgA antibody production (Table S4). Of note, the overall median IgG MFIs of the postvaccination sera (median: 1926.0, IQR: 1033.0–2638.5) were 7.9 times higher compared to the IgA signal intensity (median: 243.6, IQR: 164.3–500.3, p < 0.0001).
3.3 Differences in antibody responses to influenza A infection versus vaccination
3.3.1 Influenza A/H1N1pdm infection versus vaccination
A visual representation of microarray responses (H1N1 natural infection and vaccination) is shown in Figure 2, which demonstrates a clear distinction between patients with natural infection versus vaccination. In order to determine differences in fold changes of IgG antigen reactivity between influenza A/H1N1-infected and vaccinated SOT recipients, SAM analysis was performed, which revealed 13 A/H1N1 antigen reactivities to be significantly (q-values <0.01) different in the influenza A/H1N1-infected group versus the vaccinated group. Hierarchical clustering of these antigens revealed that patients with high IgG responses were derived exclusively from the infected group (i.e., all 13 differentially expressed antigen reactivities were higher in the natural infection group vs. the vaccine group) (Figure 2). Similarly, SAM analysis identified 11 A/H1N1 antigen IgA reactivities, which were significantly (q-values <0.01) different in the influenza A/H1N1-infected group compared to the vaccine group (Figure S4). Clustering of the IgA reactivities was less clear. However, two main clusters were detected and most (11/13) patients in the cluster with high IgA responses represented patients with A/H1N1 infection.
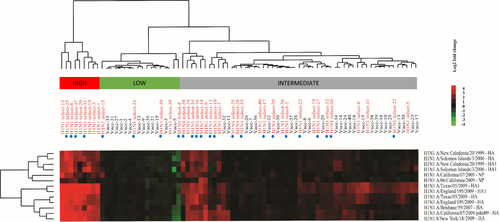
Patients who were naturally infected despite having had an influenza vaccine within the same influenza season (24/40 H1N1-infected patients) did not show a differing response pattern than unvaccinated patients (Figures 2 and S4).
3.3.2 Influenza A/H3N2 infection versus vaccination
A visual representation of microarray responses (H3N2 natural infection and vaccination) is shown in Figure 3, which again shows a clear distinction between patients with infection versus vaccination. SAM analysis identified 18 significantly different A/H3N2 IgG antigen reactivities among A/H3N2-infected and vaccinated SOT patients. Hierarchical clustering demonstrated that infected patients represented the vast majority (25/27; 92.6%) of the patients who clustered in the high responder group, while vaccinated patients clustered primarily in the low responder group (Figure 3). Similarly, for IgA, compared to vaccination, influenza A/H3N2 infection resulted in significantly (q-values <0.01) higher fold changes in reactivity against 17 antigens (Figure S5). Again, all these antigens were either complete HA proteins or HA peptides.
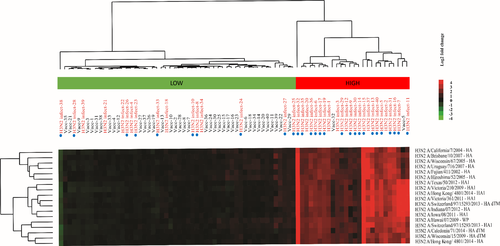
Patients who were naturally infected despite having had an influenza vaccine within the same influenza season (24/40 H1N1 infected patients) did not show a differing response pattern than unvaccinated patients (Figures 3 and S5).
3.4 Seroconversion for H1N1 A/California/07/2009 hemagglutinin antibodies by HIA and microarray
In order to determine the concordance between seroconversion (≥4-fold rise from prevaccination to postvaccination in vaccinated patients or onset of illness vs. convalescence in infected patients) and ≥4-fold rise in MFI-B, all sera underwent HIA for H1N1 A/California/07/2009. This specific antigen was chosen since it was circulating during the 2010–2015 influenza seasons. It was also included in the 2016–2017 vaccine. Using this antigen, the inter-test reliability of HIA and microarray for seroconversion was 85.0% (concordant results for 102/120 tests) (Table S5).
4 DISCUSSION
Using custom antigen microarrays, we profiled the diversity of antibody responses in immunosuppressed transplant cohorts of patients infected with influenza A/H1N1 and A/H3N2 and compared them to vaccinated transplant recipients. This led to a number of novel findings. We found that natural infection elicits a relatively robust and diverse antibody response. These antibodies are primarily directed toward the antigens of the corresponding influenza A subtype and mainly to either the whole HA protein or a subunit. However, a proportion of patients also showed an increase in antibodies to conserved antigens (such as M1, NP, and NS1) as well as to heterosubtypic antigens (i.e., antigens from other strains of influenza). We also showed that the diversity of antibody response is substantially greater in infected rather than vaccinated patients. This was evident in the absolute numbers of antigens that elicited an overall response as well as in a comparative clustering analysis of individual antigen reactivities in vaccinated versus infected patients. We believe this provides unique insight into antigen responsiveness in the setting of immunosuppression, and lays a better foundation to develop strategies to improve vaccine immunogenicity in this patient population.
There are limited data on the diversity of antibody responses to influenza in transplant patients. However, studies in immunocompetent patients suggest that the majority of antibodies induced by natural influenza infection will target HA proteins and that there will be lower-level responses to NA and internal, more conserved, viral proteins.30 The breadth of the response to HAs induced by influenza infection mainly depends on the exposure and vaccination history of the infected individual. This phenomenon, also known as the original antigenic sin, describes the increases in antibody titers to historic strains that can occur after immunization or infection with a new influenza strain.30-32 This may explain why we found some significant antibody responses to older, currently not circulating HA proteins included in our assay. Antibodies toward the NA protein are also induced in healthy individuals by natural infection.30 However, NA antibody responses are typically lower compared to HA responses. In addition, N1 NA proteins seem to be less immunogenic than N2 NA proteins.33 In agreement with these findings, we detected anti-N2 NA immune responses in influenza A/H3N2-infected SOT recipients, but we did not find significant immune responses against N1 NAs in our A/H1N1-infected patients. Antibodies to NP have also been previously reported after natural infection in healthy individuals.34, 35 Consistent with the findings in immunocompetent persons, we also detected anti-M1 antibodies after natural IAV infection in our immunocompromised population.36, 37 We also noted that in a small minority of patients, MFI-B values decreased from the first to second sample. It is possible that depending on symptom onset, we may have missed the peak of the antibody response. Alternatively, augmentation of immunosuppression between the first and second serum collections may have had some impact on a small subset of patients.
Split virus vaccines, such as the vaccine used in our study, are manufactured using inactivated influenza viruses, which are treated with a detergent and further purified. Only the content of the HA proteins in these vaccines is standardized, and the content of the other proteins is likely to depend on the specific vaccine or even the batch of the vaccine, but such proteins are present.30 Therefore, antibodies against non-HA influenza proteins can also be induced by vaccination. In the immunocompetent population, antibody responses against M1, NP, and NA proteins have been found thus far.34, 38, 39 However, compared with natural infection, the response induced by vaccination with a split virus vaccine is relatively narrow in immunocompetent persons.30 In our study, we can confirm that these findings also apply for the immunocompromised population. But we would like to highlight that influenza vaccination in transplant recipients usually generates lower antigen-specific antibody responses in comparison to the general population due to exogenous immunosuppression. Despite this, there are no antibody cutoffs that clearly define protection from disease, although a titer of 1:40 is commonly used for vaccine-associated seroprotection. Although we find the antibody titers as noted by microarray to be relatively lower for vaccinated patients, this does not imply lack of clinical protection. We have previously shown that in a multicenter cohort of transplant patients infected with influenza, prior vaccination in the same influenza season protects patients from severe illness.1 In addition, the specific vaccine used in the study was the standard-dose (15 µg) trivalent influenza vaccine. A previous randomized trial showed that high-dose influenza vaccine (60 µg/antigen) elicited a significantly greater humoral response in organ transplant recipients versus standard-dose vaccine.40 A number of different approaches have the potential to improve and broaden influenza vaccine responses in transplant recipients including high-dose vaccines, MF59-adjuvanted vaccine, and recombinant influenza vaccines. The ability of these vaccines to elicit a greater breadth of antibodies as detected by antigen microarrays will be investigated in future studies.
Another novel aspect of this study was the measurement of IgA responses. Indeed, we found that a serum IgA antibody response is induced upon influenza A infection and also with vaccination. The responses generally correlated with IgG responses in terms of antigen reactivity. In human serum, IgG constitutes approximately 75% and IgA 15% of antibody.41 Both serum IgG and serum IgA antibody responses (measured by ELISA) to natural IAV infection and to intramuscular vaccination with an inactivated IAV vaccine have been reported previously in non-immunocompromised individuals.42-44 The role of viral-specific serum IgA for protection against influenza or viral clearance is unclear. However, the results of a human influenza challenge study in healthy volunteers suggest that there is a weak inverse correlation between influenza-specific serum IgA antibody concentration and viral shedding measured by culture.44
One limitation of our study is that our analysis was that we were restricted to antigens that were commercially available. These may not have exactly matched the antigens in circulating influenza strains or vaccine strains, although we attempted to have a diversity of antigens. In addition, the antigens used in our assay were produced by several different manufacturers using varying expression systems. This may have negatively affected the detection for some of the antibody responses because the microarray technology does not allow to optimize the assay for each individual antigen. Another limitation is that there is no specific cut-off MFI that implies protection. We addressed this by comparing fold changes in MFI-B instead of focusing on absolute values.45 It is possible that conformational epitopes may not be properly displayed for antibody binding; however, studies using the same platform for autoimmune antigens have shown correlation between traditional ELISA and antigen microarrays.24, 46 We also found a concordance of 85% between our microarray technique and HIA for one of the common influenza strains during the study period. The design of the study also meant that only medically attended cases of influenza were enrolled. This means that mildly symptomatic cases that may perhaps have a different antibody response profile after infection could be underrepresented. Furthermore, there was a slight overrepresentation of kidney transplant recipients in the naturally infected group compared to the vaccinated population. As the immunosuppressive regimens among the two groups were comparable, we do not believe that this had a major impact on our findings, although it is not possible to exclude the impact of more granular differences in immunosuppression. Non-transplant controls were not included in this study and this is a limitation. Therefore, study results need to be interpreted in this context.
In summary, we provide novel information profiling the diversity of antibody response to natural influenza infection and vaccination in the transplant population. We show that natural infection resulted in a substantially broader humoral immune response compared to vaccination with a split virus vaccine. Thus, vaccines with greater antigenic diversity may better mimic the immune responses elicited by natural infection. We also provide novel data on response to conserved virus antigens. A better understanding of the differences between natural infection and vaccination will likely be helpful to design better vaccines for immunocompromised patients.
ACKNOWLEDGMENTS
C.H. was supported by an early Postdoc. Mobility grant from the Swiss National Science Foundation (P2BEP3_175265). A.L. was supported by an advanced Postdoc. Mobility grant from the Swiss National Science Foundation (P300PB_171603).
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. D.K. has received research grants from Roche and GSK, and honoraria from GSK, Roche, Sanofi, and Merck. A.H. has received a research grant from Roche. V.H.F. has received a travel grant from Roche. The other authors have no conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS
Study Design and Concept – C.H., A.H., D.K., V.H.F.; Data Gathering – Y.N., A.G.L., E.C., S.H.H.; Analysis – C.H., V.H.F., A.C.; Writing and Review of Manuscript – All authors.
APPENDIX A1
The Influenza in Transplant Study Group
Pilar Perez-Romero, Hospital Universitario Virgen del Rocío and Biomedicine Research Institute, Seville, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Teresa Aydillo, Hospital Universitario Virgen del Rocío and Biomedicine Research Institute, Seville, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Jordi Carratala, University Hospital of Bellvitge, L’Hospitalet de Llobregat, Barcelona, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Patricia Munoz, Gregorio Marañón University Hospital, Instituto de Investigación Sanitaria Hospital Gregorio Marañón, CIBER Enfermedades Respiratorias-CIBERES, and Department of Medicine, School of Medicine, Universidad Complutense de Madrid, Madrid, Spain; Miguel Montejo, Cruces University Hospital, Vizcaya, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Francisco Lopez-Medrano, University Hospital 12 de Octubre, Instituto de Investigación Biomédica i + 12, Madrid, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Maria Carmen Farinas, University Hospital Marqués de Valdecilla, Santander, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Joan Gavalda, Vall d’Hebron University Hospital, Barcelona, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Asuncion Moreno, University Clinic Hospital, Barcelona, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Jesus Fortun, University Hospital Ramón y Cajal, Madrid, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Julian Torre-Cisneros, Maimonides Biomedical Research Institute of Cordoba, Reina Sofia University Hospital, University of Cordoba, Córdoba, Spanish Network for Research in Infectious Diseases (REIPI), Spain; Emily Blumberg, University of Pennsylvania, USA; Lara Danziger-Isakov, Cincinnati Children's Hospital, USA; Ajit Limaye, University of Washington, USA; Marilyn Levi, University of Colorado, USA; Peter Chin-Hong and Catherine Liu, University of California, San Francisco, USA; Tanvi Sharma, Boston Children's Hospital, USA; Janet Englund, Seattle Children's Hospital, USA; Gail Reid, University of Illinois, USA; Fernanda Silveira, University of Pittsburgh, USA; Shahid Husain, University of Toronto, Canada.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




