Hiding in Plain Sight—A New Look at HLA Epitopes: A Case Report
Abstract
“Epitope matching” has become a buzz word in solid organ transplantation. Its goal is to improve matching between donor and recipient, to minimize risk for antibody-mediated rejection and to reduce sensitization associated with graft failure. Current software allows identification and enumeration of amino acid sequence mismatches in the form of HLA eplets; however, “eplets” and “epitopes” are not interchangeable terms, and the understanding of what contributes to the antigenicity and immunogenicity of HLA B cell epitopes is still very limited and inadequate. In fact, we still do not know what constitutes an HLA epitope or how to define it in a clinically useful way. To allow for judicious implementation of epitope matching, it is critical to explore the full spectrum of factors that affect allorecognition. In exploring antibody-binding patterns, we have uncovered a potential tool—currently hidden in plain sight—that may shed light on some aspects of epitope characteristics.
Abbreviations
-
- AMR
-
- antibody-mediated rejection
-
- CREG
-
- crossreactive group
-
- DSA
-
- donor-specific antibody
-
- HLA-DSA
-
- donor-specific HLA antibody
-
- MFI
-
- mean fluorescence intensity
Introduction
The development of donor-specific HLA antibodies (HLA-DSAs) after transplant is associated with increased risk for antibody-mediated rejection (AMR) and poor allograft outcome. Although development of AMR can be associated at times with medication noncompliance, it is not clear why some recipients will develop donor-specific antibodies (DSAs) to their HLA-mismatched donors, whereas others will not. Over the years, research has focused on deciphering which donor–recipient HLA mismtaches can be considered acceptable 1-3.
In recent years, the term “epitope matching” was introduced, mainly as a result of the growing use of the computer algorithm HLAMatchmaker. This software allows a user-friendly comparison between donor and recipient HLA antigens and identification of the number of amino acid sequence differences between them, termed “eplets.” Multiple studies have used this software, with the highlight recently published by Nickerson and his group 4, 5. In these studies, eplet load (i.e. the overall difference in HLA class II amino acid sequences between donor and recipient) was calculated. Their data clearly demonstrated that determining eplet load is a powerful tool to risk-stratify patients with regard to their likelihood of developing de novo HLA antibodies and to individualize immunosuppression and posttransplant monitoring. These studies confirmed and advanced data published previously by other groups 6, 7.
The ability to determine eplet load, however, does not explain what contributes to the antigenicity and immunogenicity of an epitope. It is important to appreciate that HLA epitopes are physically larger than eplets. In fact, the eplet contributes an area of about one-fifth of the overall area of the epitope. Although a critical part of the epitope, an eplet on its own is most likely not sufficient to drive an immune response (for review, see Tambur and Claas 8). Duquesnoy himself published that for an HLA antibody to recognize nonself component of the HLA antigen, it has to be paired with a self component (eplet 9-11). This fact is critical because it delves into the very basics of the HLAMatchmaker software 12. The current algorithm of HLAMatchmaker starts with eliminating all self eplets to compare the mismatched sequences and to count eplet load. Given that self eplets are likely required to form the functional epitope, removing self eplets may yield misleading conclusions.
The quest to decipher what constitutes an HLA B cell epitope may prove to be quite elusive. In many cases, especially when the recipient is highly sensitized, HLAMatchmaker fails to give a clear response. Multiple assumptions are required to provide a hypothetical string of eplets; otherwise, the result shows no immunogenic eplets at all. Even when the software suggests an eplet, multiple alleles carrying the same eplet will show different levels of reactivity 13. Importantly, currently many “epitope” assignments are performed in a retrospective fashion whereby modification of interpretation needs to be introduced to fit current paradigms. In fact, current tools do not provide mechanisms to predict which eplet mismatch may be immunogenic in a specific donor–recipient combination. Interestingly, new understanding of HLA epitopes may come from analyzing sera from highly sensitized patients, for whom antibody specificity was historically considered more difficult to interpret. Data emerging from titration studies illustrates antibody grouping patterns outside of current convention and provides a new avenue for investigating HLA epitopes. Two cases are presented to demonstrate this point.
Patients and Methods
Patient 1 is a white women 63 years old, with 0% panel reactive antibody documented by FlowPRA in three consecutive serum samples. She received a living donor kidney transplant from an unrelated white woman 25 years old. The patient developed strong DSAs to both donor DQ antigens within about 2 weeks after transplant, indicating a likely memory response that was quiescent during her pretransplant evaluation.
The second case presents a black man 50 years old who was sensitized prior to transplant, with no historic or current DSAs at the time of transplant. The patient developed de novo DSAs ≈2 years after transplant.
HLA typing was performed using LABType One Lambda, A Thermo Fisher Scientific Brand, Canoga Park, CA with ambiguity resolution. HLA antibody testing was performed using LABScreen Single Antigen antibody-detection tests (One Lambda). Both methodologies followed the manufacturer's guidelines, as published previously 14.
Results
In patient 1, initial testing after transplant indicated a strong inhibitory effect (neat mean fluorescence intensity [MFI] values, left hand side of Figure 1), namely, neat MFI values did not correlate with antibody strength. Peak MFI values (Figure 1) indicated that 25 of 28 DQ-coated beads were positive, with MFI values of ≥8000. Moreover, dilution studies revealed that different antibodies have diverse titration patterns and that inhibitory factors in the serum have distinct effects on different antibodies (Figure 1).
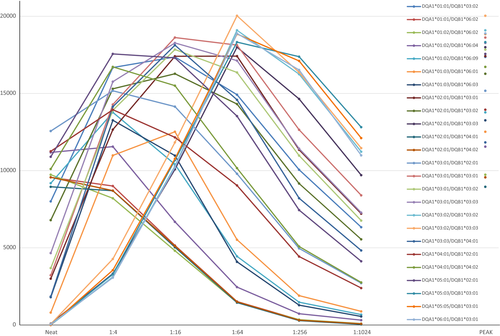
The specific clusters are illustrated separately in Figure 2(A–E). Some of those patterns seem to follow current conventions of HLA-DQ antibody nomenclature. Specifically, Figure 2(A) shows two HLA-DQ alleles exhibiting strong inhibition in the undiluted sample, with peak MFI values reached around a titer of 1:4–1:16 (MFI values of ≈13 000). The antibodies titer out (become negative) at a dilution of ≈1:256. The two alleles are DQA1*01:03/DQB1*06:01 and DQA1*01:03/DQB1*06:03, suggesting a shared target/epitope encoded by DQA1*01:03.
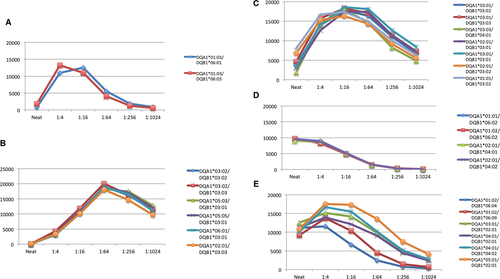
Figure 2(B) shows another seemingly expected cluster of HLA-DQ alleles, including DQA1*03:02/DQB1*03:02, DQA1*03:02/DQB1*03:03, DQA1*05:03/DQB1*03:01, DQA1*05:05/DQB1*03:01, DQA1*06:01/DQB1*03:01 and DQA1*02:01/DQB1*03:03; all are alleles of the serologic DQ3 specificity. This group of alleles starts at low MFI values and reaches peak levels at a titer of 1:64 (MFI values 17 000–20 000). These antibodies remain strong even at a dilution of 1:1024. The almost identical dilution patterns and the ability to presume one familiar target for these antibodies again suggests a common epitope shared by the targets.
Figure 2(C) shows another pattern of dilutions that is different from Figure 2(B) despite the fact that, by serologic nomenclature, those antibodies also recognize members of the DQ3 group (including DQ7, DQ8, and DQ9 but with different DQα chains). Interestingly, the World Health Organization nomenclature committee and the National Marrow Donor Program currently recognize both DQA1*03:01:01 and DQA1*03:02 as identical for clinical purposes, and both are referred to as DQA1*03:01:01G. This is a reflection of current thinking, assuming that because the amino acid differences between these two alleles are outside of the peptide-binding site (exon 2), they can be considered a nonsynonymous polymorphism. Nonetheless, two DQ molecules that share the same DQB1*03:02 but are combined with the two different DQA1 alleles (DQA1*03:01:01/DQB1*03:02 and DQA1*03:02/DQB1*03:02) show different dilution patterns (Figures 2B and C, respectively). This observation raises doubts regarding the original assumption about grouping alleles within a “G” category. Although hypothetical, this example suggests that changes outside of exon 2 can lead to differences in the overall HLA protein conformation, with ramifications for both antibody binding and T cell receptor binding.
Figure 2(D) groups four alleles with identical dilution patterns but two very distinct serologic specificities. Two are serologic DQ6, and the other two are serologic DQ4. The likelihood of such an observation without physiologic similarities among the four alleles seems very remote. Figure 2(E) compiles the remaining positive responses. Although the pattern is somewhat similar, the differences in MFI values along the titration curve indicate that those are likely different groups of epitope-recognizing antibodies.
For this particular patient, both HLA-DQ DSAs clearly elicited an immune response. DQA1*05:03/DQB1*03:01 is present in Figure 2(B) and likely elicited a response to all other HLA-DQ alleles present in this group. DQA1*03:01/DQB1*03:02 is present is Figure 2(C) and likely triggered a response to all other alleles in that cluster. The source for the other antibody responses is likely associated with prior sensitization (as indicated by the very rapid antibody production response after transplant).
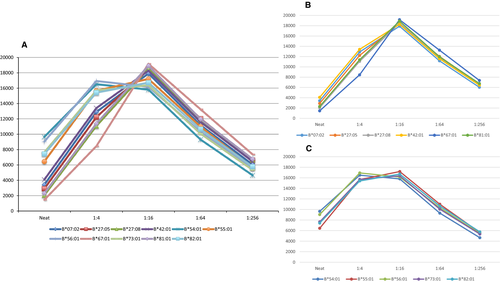
Patient 2 illustrates that this phenomenon of seemingly similar antibodies having different dilution patterns is not restricted to HLA-DQ or to class II antibodies. Figure 3(A) shows a patient that developed antibodies to what can be considered a HLA-B7 crossreactive group (CREG). All antibodies in this example exhibit inhibitory effects, and all have peak MFI values ranging between 16 000 and 19 000 MFI units. Although there are several permutations of B7 CREG presentations (including some but not all members of the CREG), when a antibodies with similar characteristics have comparable MFI values, the tendency is to assume that all react with the same target; however, two clear patterns of dilution are apparent when looking at the configurations associated with titration studies (Figures 3B and C). Consequently, we believe that what could have been interpreted as antibodies to a single epitope is in fact a result of two different antibodies recognizing two different epitopes.
Discussion
Compelling evidence was recently submitted to show that factors other than the mere eplet/amino acid sequence are involved in the definition of HLA epitopes. Kosmoliaptsis et al elegantly showed that the immunogenicity and antigenicity of HLA molecules are dependent on electrostatic potential and physiochemical properties of the molecule. Using such an approach, they were able to predict humoral alloresponses 15-17. Zachary's group analyzed the rate of antibody production among mirror-image pairs (situations in which A is recipient and B is donor and then the reverse). The frequency of antibody production in both cases should be similar if immunogenicity is based purely on the number of eplet mismatches; however, in some pairings, antibody production was skewed to one direction 18, nulling the base hypothesis.
Dilution studies provide more dynamic surveillance of antibody behavior than the static single measure of the routine Single Antigen Luminex assay. We demonstrated that antibodies may be grouped in clusters outside of current nomenclature conventions, potentially providing “out-of-the-box” tools to decipher antibody reactivity. Titration studies are far from being a new approach to study characteristics of antibodies. In fact, titers have been used since the inception of the field of immunology to better describe immune responses; therefore, it is even more compelling to reintroduce well-defined techniques with today's knowledge of HLA antigen and antibody specificities. We hypothesize that antibody clusters, as defined by dilution patterns, can provide clues about shared antibody targets and thus the physiologic epitope. In that respect, allowing inhibitory factors to prevail (what is referred to by many as “prozone”) can in fact shed some light into epitope analysis.
Berth 19 correctly remarked that the exact causes for the interference effects observed in Luminex single-antigen assays are not fully understood. They are likely multifactorial, including complement interference, IgM interference, polyclonallity and the true prozone phenomenon (defined as oversaturation of the capture and detection antibodies). An additional mechanism may be a competition by different antibodies “fighting” to bind a respective but somewhat overlapping target on the surface of the same antigen. Although dilution patterns can result from different causes, the data in support of such hypothesis are quite compelling. Additional work is required to provide definitive proof, for example, absorption/elution studies; follow-up studies on data similar to that reported by the Nickerson group 4 comparing patients that developed antibodies with those who did not despite similar donor HLA mismatching; and analysis of proposed epitopes with regard to electrostatic potential, similar to the work reported by Kosmoliaptsis et al 16.
The ramifications of epitope matching encompass not only better matching for the current transplant but, more important, minimization of the risk of developing de novo HLA-DSAs after transplant. Consequently, the risk of AMR should be reduced, lowering the probability of converting patients to a highly sensitized state if the need for retransplant arises. Deciphering the enigma of HLA B cell epitopes is likely to substantially transform the field of solid organ transplantation. To judiciously pursue our understanding of HLA epitopes, it is critical to make the distinction between an eplet and an epitope and to use the appropriate terminology carefully. The factors that contribute to defining epitope antigenicity and immunogenicity should be decrypted, and we may need to consider a new notation system to account for these findings. The ramifications of the observations presented are not yet clear and require additional investigation; however, if we assume that the observed patterns are not random, then there might be a biological reason behind it. We submit that dilution patterns can provide a new tool and new insight into the study of HLA epitopes, as viewed by HLA antibodies. In other words, it may support the study of “epitopology” (as coined by Frans Claas).
Disclosure
The author of this manuscript has no conflicts of interest to disclose as described by the American Journal of Transplantation.




