Optimisation of techniques for quantification of Botrytis cinerea in grape berries and receptacles by quantitative polymerase chain reaction
Abstract
Background and Aims
Bunch rot symptoms can appear weeks after the infection of grape flowers by Botrytis cinerea. Quantitative polymerase chain reaction (qPCR) detects changes in the DNA mass of a target organism and is a potential tool for studying quiescent infections. The aim was to optimise a duplex qPCR to quantify B. cinerea DNA in the background of endogenous Vitis vinifera DNA.
Methods and Results
Three DNA extraction techniques and three probe sets were compared. The optimised qPCR using the Bc3 probe set was 1000-fold more sensitive than other probe sets, with a threshold cycle value of <33 for as little as 1 picogram of B. cinerea DNA. The duplex assay successfully detected an increasing amount of B. cinerea DNA when mixed with V. vinifera DNA or of B. cinerea conidia when added to grape receptacles.
Conclusions
Duplex assays quantifying B. cinerea DNA in the background of endogenous grape DNA were efficient and sensitive, with calculation of a pathogen coefficient allowing comparison of results among assays.
Significance of the Study
The results demonstrate the potential to monitor symptomless, quiescent infections and to investigate the consequence of an intervention (e.g. a fungicide treatment) before disease symptoms are visible.
Introduction
Botrytis cinerea Pers. is a ubiquitous fungal pathogen that causes grey mould on a large number of economically important agricultural and horticultural crops. In grapevine, B. cinerea causes grapevine bunch rot and grey mould, resulting in loss of yield and quality of grapes. The first opportunity for the pathogen to infect fruit is at flowering, which is followed by a period of latency, during which the pathogen is present inside the berry without exhibiting any disease symptoms until berries begin to ripen (McClellan and Hewitt 1973, Nair et al. 1995). Flower infections are believed to be an important stage in the epidemiology of B. cinerea in grape (Nair et al. 1995, Keller et al. 2003, Viret et al. 2004). Nair et al. (1995) reported a direct quantitative relationship between flower infections and the incidence of grey mould at harvest. Keller et al. (2003) reported that the most likely site of infection at flowering is the receptacle area, which has also been supported by preferential isolation of the pathogen from this region (Holz et al. 2003) and electron microscopy observations (Viret et al. 2004). A direct polymerase chain reaction (PCR) assay on pea-sized berries and receptacles revealed that B. cinerea was present only in the receptacles (Gindro et al. 2005). Conventional PCR assays, however, do not provide precise quantification. Recently, the introduction of real-time quantitative PCR (qPCR) has provided a mechanism to simultaneously detect and quantify DNA of specific organisms (Angelini et al. 2007, Atallah et al. 2007, Boonham et al. 2008, Brunner et al. 2009, Carisse et al. 2009, Celik et al. 2009, Mideros et al. 2009, Park et al. 2011). Comparisons among samples are made based on the threshold cycle (Ct) when each sample exceeds a fluorescence threshold. Advantages of qPCR include its generally high sensitivity and specificity, which make this technique suitable for multiple uses in biological research and potentially for applications that inform disease management decisions.
Botrytis cinerea detection by qPCR has been reported by several authors. For instance, Suarez et al. (2005) tested several target genes for B. cinerea detection in Pelargonium sp. Similarly, Cadle-Davidson (2008) developed a species-specific primer for a qPCR protocol based on a hydrolysis probe. Colonisation of grape berries by B. cinerea was monitored by this method at four growth stages, from pea-sized berries to fruit ripeness. The B. cinerea detection rate, however, at the pea-sized berry stage was low, likely because of poor DNA purity and/or assay inefficiency that resulted in reduced sensitivity. Although PCR-based detection methods are rapid, sensitive and reliable compared with that of conventional light microscopy analysis, and often more sensitive than that of immunodetection techniques (Celik et al. 2009, Diguta et al. 2010), organic compounds such as polyphenols and polysaccharides present within plant tissues can inhibit or enhance PCR amplification (Wilson 1997, Varma et al. 2007), causing a false negative reaction or delayed amplification. Thus, preparation of high-quality DNA could be a key factor in development of robust qPCR methods.
This study focused on improving current qPCR technology for the detection of B. cinerea in grapevine. The first objective was to compare the yield and purity of Vitis vinifera DNA resulting from application of three different DNA extraction methods. The second objective was to compare the performance of primer and probe sets for quantification of B. cinerea DNA. The third objective was to develop a duplex qPCR to quantify V. vinifera and B. cinerea DNA and to check for false negative results. The duplex qPCR was then applied to grape receptacles spiked with B. cinerea conidia to investigate the efficiency of extraction and detection from living tissues and application of a pathogen coefficient (PC) for comparisons of results from different assays.
Materials and methods
Fungal and plant materials
The single-spore isolate W01 of B. cinerea used in this study was collected in 2010 from a vineyard of the Vitis interspecific hybrid cv. Vignoles located at the New York State Agricultural Experiment Station near Geneva, NY using the technique that previously described by Saito et al. (2009). The isolate was maintained on potato dextrose agar (Difco, Franklin Lakes, NJ, USA) and fungal mycelium for DNA extraction was obtained by scraping the surface of 7–10-day-old cultures. Extraction of DNA is described later.
Plant material was collected from pea-sized berries of the ‘Pixie’ mutant of V. vinifera cv. Pinot Meunier, grown from green cuttings in a growth chamber. Berries with receptacles were harvested. The receptacles were separated from the berries using sterile secateurs to obtain the desired weight of each tissue type and then frozen in liquid nitrogen. Samples were stored at −80°C until use. The absence of internal latent B. cinerea in the plant material was confirmed by plating some berries and receptacles on potato dextrose agar medium at 25°C for a week, on which no growth was detected.
Yield and purity of DNA
Berry or receptacle tissue (100 mg) previously frozen and stored at −80°C was placed in a 2-mL centrifuge tube with a 5-mm stainless-steel bead (OPS Diagnostics, Lebanon, NJ, USA) and ground by shaking in a TissueLyser (Retsch Inc., Newtown, PA, USA) for 1 min at 30 cycles/s. A preliminary experiment indicated a yield of DNA with the TissueLyser grinding method similar or higher than that obtained by grinding tissues to a fine powder in liquid nitrogen with a mortar and pestle (data not presented). TissueLyser was also selected because it enables a higher sample throughput (up to 48 at a time) than manual grinding.
Three methods were compared for the extraction of DNA from young grape berries. The first (I) was a slight modification of that described by Cadle-Davidson (2008), substituting a 2-mL centrifuge tube for a 96-well plate. The second (II) was described by Celik et al. (2009), and the third (III) was a slight modification of that described by Saito et al. (2009) and was used for both plant and fungal DNA extraction. Briefly, the ground tissue was transferred into a 1.5-mL microcentrifuge tube and was mixed with 500 μL of cetyltrimethylammonium bromide (CTAB) extraction buffer (2% CTAB, 100 mM Tris-HCl pH 8.0, 20 mM ethylenediaminetetraacetic acid (EDTA), 1.4 M NaCl, 1% polyvinylpyrrolidone). The mixture was then shaken vigorously with a vortex mixer and heated for 15 min at 65°C. Five-hundred microlitres of chloroform-isoamyl alcohol (24:1, v:v) was added. After further vigorous shaking, the tube was centrifuged at 12 000 rpm for 5 min, and the supernatant was transferred to a new microcentrifuge tube. The chloroform-isoamyl alcohol centrifugation and supernatant transfer were repeated. The supernatant was transferred to a new microcentrifuge tube, and a 65°C solution of 10% CTAB with 0.7 M NaCl was added at a rate of 1:10 (v/v). A third chloroform-isoamyl alcohol extraction and centrifugation was performed, and the resulting supernatant was transferred to a new microcentrifuge tube, to which was added an equal volume of cold (approximately 0°C) isopropanol and a 10% volume of 3 M sodium acetate, followed by centrifugation at 12 000 rpm for 5 min at 4°C. After centrifugation, the pellet was washed with 70% (v/v) ethanol, air dried and resuspended in 100 μL of sterile distilled water.
Each DNA extraction method was applied to two independently collected tissue samples, and this process was repeated three times resulting in a total of six DNA solutions per extraction method. The yield and purity of extracted DNA were determined spectrophotometrically by a NanoDrop instrument (NanoDrop Technologies, Wilmington, DE, USA) and normalised to the desired concentration.
Absorbance ratios A260/A280 and A260/A230 were calculated to indicate contamination by protein and by polyphenols and carbohydrates, respectively (Manning 1991, Varma et al. 2007). A ratio of 1.8–2.0 for A260/A280 and a ratio >1.8 for A260/A230 indicate DNA with little contamination (Yeates et al. 1998).
The purity of each DNA solution was determined further by testing amplification of V. vinifera resveratrol synthase gene I, using the probe set reported by Valsesia et al. (2005) with the fluorescent reporter 6-carboxyfluorescein (FAM) being substituted for hexachlorofluorescein (HEX). The sequences are: Res F: 5′-CGA GGA ATT TAG AAA CGC TCA AC-3′; Res R: 5′-GCT GTG CCA ATG GCT AGG A-3′; Res P-HEX: 5′-HEX-TGC CAA GGG TCC GGC CAC C-BHQ-2-3′. Vitis vinifera DNA (10 ng/μL) was serially diluted (from 10 ng/Ll to 0.01 ng/μL) in sterile water. The qPCR was conducted using a 10 μL total volume in a well of a 96-well PCR plate which contained 1 unit of HS Taq (Takara Ex Taq DNA Polymerase Hot Start Version; Takara Bio Inc., Shiga, Japan), 1 μL of 10 × Ex Taq Buffer, 0.4 μL of deoxynucleotide triphosphate (dNTP) mixture (2.5 mM each), 1 μL DNA (desired concentration), 150 nM probe and 500 nM each primer. After incubation at 95°C for 3 min, 40 cycles of a two-step amplification were run at 95°C for 15 s and 60°C for 45 s using a Bio-Rad iQ cycler system (Bio-Rad, Hercules, CA, USA). Each DNA sample was assayed twice with qPCR, resulting in two standard curves. The slopes of the efficiency equation of the two standard curves were subjected to an analysis of covariance (ANCOVA) using PROC GLM in SAS (v. 9.3; SAS Institute, Cary, NC, USA). If there was no significant difference between the two slopes, all data were pooled to create one standard curve.
Comparison of probe sets for B. cinerea
The sensitivity of three probe sets for quantifying B. cinerea DNA was examined using B. cinerea DNA (10 ng/μL) serially diluted (tenfold dilutions from 10 ng/μL to 0.001 ng/μL) in sterile water. Two of the probe sets have been reported previously: one (Bc3) targeting an intergenic spacer region between 28S and 18S genes (Suarez et al. 2005) and another (Bc Taq) targeting a non-coding sequence adjacent to a hypothetical protein (Cadle-Davidson 2008). To test whether predicted hairpins and dimers in the Bc Taq assay could reduce sensitivity, a third probe set (UTAS) was independently developed at the University of Tasmania to target the same intergenic region as Bc Taq. The sequences are as follow: (i) for Bc3, Bc3F: 5′-GCT GTA ATT TCA ATG TGC AGA ATC C-3′; Bc3R: 5′-GGA GCA ACA ATT AAT CGC ATT TC-3′; and Bc3P: 5′-6-Fam-TCA CCT TGC AAT GAG TGG-BHQ-1-3′; (ii) for Bc Taq, BcTaq424f: 5′-GCT TCC CCC GTA TCG AAG A-3′; BcTaq491r: 5′-CGA ACG GCC AGG TCA TCT-3′; and BcFamP: 5′-6-Fam-CCC TAG ATT TGA TTT TAC CCT TCG CGT GG-BHQ-1-3′; and (iii) for UTAS, UTAS-F: 5′-GGA CTT GGA CAT GGA TAC-3′; UTAS-R: 5′-ACA ATC AAA GAC CAG AGG-3′; and UTAS-P: 5′-6-Fam-CAC TCG CAC CTA ATT CGT CAA CG-BHQ-1-3′. Reaction mixtures and cycler conditions were the same as those described previously. For each probe set, the qPCR experiment was performed twice with three replicates of each dilution series. All six replicates for each probe sets were pooled if there was no significant difference between the slopes according to ANCOVA as described earlier.
Comparison of HS Taq and a pre-assembled reaction mixture
Pre-assembled reaction mixtures purchased ‘off the shelf’ are convenient; this convenience, however, may correspond to a higher cost per reaction than those prepared from individual reagents. The HS Taq mixture was compared with a widely used, pre-assembled mixture (iQ Supermix, Bio-Rad, Hercules, CA, USA) to check if this cheaper mixture could be used routinely without any loss in the quality of the qPCR. For qPCR with iQ Supermix, 5 μL 2 × iQ Supermix was added to the 10 μL reaction, instead of HS Taq, Ex Taq buffer and dNTP mixture. Cycling conditions were as previously mentioned, and qPCR assays were performed twice.
Effect of grape DNA on amplification of fungal DNA and PC
Concurrent amplification of B. cinerea and the V. vinifera target DNAs in a duplex qPCR was tested using the V. vinifera probe set (Valsesia et al. 2005) and the Bc3 probe set. Botrytis cinerea DNA (10 ng/μL) was serially diluted in sterile water as before. One μL of each B. cinerea DNA dilution was mixed with 1 μL of V. vinifera DNA (20 ng/μL), producing a tenfold dilution series (from 1:2 to 1:20 000 w/w pathogen : grape DNA). This dilution series was established during preparation of the duplex qPCR mixtures as follows: a 10 μL reaction volume contained 1 unit of HS Taq, 1 μL of 10 × Ex Taq Buffer, 0.5 μL of dNTP mixture (2.5 mM each), 1 μL of each normalised grape and pathogen (diluted) DNA, 150 nM V. vinifera probe, 150 nM B. cinerea (Bc3) probe, 100 nM each V. vinifera primer and 500 nM each B. cinerea (Bc3) primer. The mixtures were amplified under the same conditions as described earlier. The qPCR experiment was performed twice with three replicates for each dilution of B. cinerea DNA in V. vinifera DNA. Data for all six replicates for each dilution were pooled if there was no significant difference between the slopes according to ANCOVA as described earlier.
For each duplex assay, the PC is the Ct number for the V. vinifera gene, amplified from a constant amount of grape DNA, divided by that for B. cinerea DNA, which is likely to vary among samples. In short, PC = CtV. vinifera/CtB. cinerea (Valsesia et al. 2005).
Quantification of B. cinerea DNA in the receptacle
Conidia were harvested by flooding 2-week-old cultures of B. cinerea on 9-cm diameter petri dishes with sterile distilled water and suspended in sterile distilled water. The resultant conidial suspension was filtered through autoclaved gauze; the conidial concentration was then quantified microscopically with a haemocytometer and diluted to the desired concentration described later. Conidial suspensions were prepared immediately prior to inoculation and placed on ice until use.
One 2 μL droplet of a conidial suspension, providing a total of either 10, 50, 250 or 1250 conidia, was inoculated onto 50 mg of the receptacle dissected from Pixie berries (derived from approximately three individual berries), and the plant material was then immediately ground as described earlier. DNA was extracted using method (III) for application of the duplex assay and calculation of the PC as described earlier. This test was performed twice.
Results and discussion
Yield and purity of grape DNA
Extraction methods (I) and (III) yielded more V. vinifera berry DNA than method (II) (Table 1). Extraction method (III) applied to berry or receptacle consistently provided DNA with A260/A280 and A260/A230 ratios meeting criteria for high-purity DNA (Table 1). DNA extracted by method (I) did not amplify the target gene when 10 ng DNA was used, suggesting that polyphenolic or carbohydrate contaminants inhibited the PCR (A260/A230 = 0.90) (Table 1). In contrast, DNA extracted from either berry or receptacle by method (III) yielded standard curves with a high correlation between Ct values and the different dilutions of grape DNA (Table 1 and Figure 1). There was no significant difference between the berry and receptacle standard curves according to ANCOVA analysis (P = 0.0983).
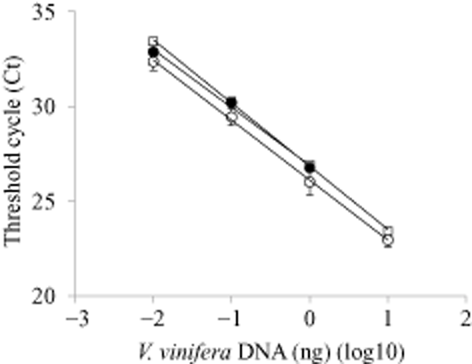
Standard curves for detection of Vitis vinifera resveratrol synthase gene I from genomic DNA extracted by two methods (I and III) from grape berries or the receptacle. Threshold cycles (Cts) were plotted against the logarithm of the mass (ng) of a genomic DNA standard, with each data point representing the mean of six replications from two assays and error bars representing one standard deviation. Method I, berry (•); Method III, berry (○); Method III, receptacle (□). Regression equations and R2 values are shown in Table 1.
| DNA extraction method† | Grapevine tissue | Total yield (μg/100 mg) (mean ± SE) | Absorbance ratio | Regression curve‡ | R2 | |
|---|---|---|---|---|---|---|
| A260/A280 (mean ± SE) | A260/A230 (mean ± SE) | |||||
| I | berry | 2.09 ± 1.7 | 1.77 ± 0.02 | 0.90 ± 0.01 | y = −3.06x + 26.9 | 0.995 |
| II | berry | 1.8 ± 0.5 | 1.69 ± 0.10 | 0.25 ± 0.10 | ND | ND |
| III | berry | 24.7 ± 1.9 | 1.98 ± 0.01 | 2.08 ± 0.06 | y = −3.17x + 26.1 | 0.999 |
| III | receptacle | 29.4 ± 4.0 | 1.87 ± 0.02 | 1.95 ± 0.15 | y = −3.34x + 26.8 | 0.999 |
- †Three different DNA extraction methods were used, as described in the methods: I, Cadle-Davidson (2008); II, Celik et al. (2009); and III, modified from Saito et al. (2009). Samples were ground by shaking in TissueLyser with a 5-mm stainless-steel bead for 1 min at 30 cycles/s. ‡Regression curve equations and R2 value for detection of Vitis vinifera resveratrol synthase gene I. Standard curves are shown in Figure 1. ND, no data; SE, standard error.
Comparison of probe sets for B. cinerea
The standard curves obtained showed an equally, extremely strong log-linear response between the Ct value and fungal DNA quantity for all three probe sets (R2 = 0.997–0.999) (Figure 2). At a given Ct, however, the Bc3 probe set was nearly 1000-fold more sensitive than the Bc Taq or UTAS probe sets (Figure 2). Similarly, when the reaction was initiated with 0.001 ng of the purified B. cinerea DNA, the Bc3 Ct value was less than 33 while the other two probe sets failed to amplify within 40 cycles at this dilution (Figure 2).
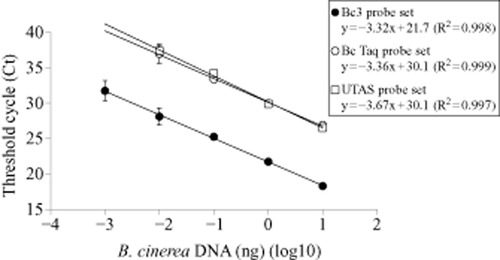
Standard curve of Botrytis cinerea amplicon obtained with three different probe sets: Bc3 (•), Bc Taq (○) and UTAS (□). Threshold cycles (Cts) were plotted against the logarithm of the mass (ng) of a genomic DNA standard. Each data point represents the mean of six replications from two assays. Error bars represent one standard deviation.
Previously, Diguta et al. (2010) developed B. cinerea detection by qPCR using the same primer Bc3, but they could not detect a low concentration of purified B. cinerea DNA (less than 10 pg). They ascribed this suboptimal sensitivity of detection to their use of SYBR green method (Diguta et al. 2010). Relative to their results, the sensitivity of detecting B. cinerea was at least an order of magnitude improved in both the current study and that of Suarez et al. (2005), both of which used hydrolysis probes.
The higher sensitivity of the Bc3 probe set can be explained by the fact it targets rDNA, which has a high copy number. While the high copy number makes rDNA a desirable target for sensitive qPCR detection of a wide array of plant pathogens, there are potential shortfalls of targeting rDNA for this purpose: (i) without a known copy number, quantification remains relative and not absolute; and (ii) copy number might vary among isolates of B. cinerea – although this has not been reported for B. cinerea or any fungus (Lievens and Thomma 2007). This variability, if present, would affect even relative qPCR-based estimates if copy number was not consistent between samples. Thus, each experimental design should consider the tradeoffs between targeting a multicopy gene for improved sensitivity or a single-copy gene that may suffer from false negative errors when the extent of plant tissue colonisation is low. Given the low level of natural infection by B. cinerea in developing grape berries (Cadle-Davidson 2008), the increased sensitivity of the Bc3 probe set should be useful for applications requiring detection of the fungus.
Application of either HS Taq or iQ Supermix reaction mixtures resulted in similar standard curves with high R2 values (y = −3.29x + 23.6, R2 = 0.998 for HS Taq and y = −3.22x + 23.4, R2 = 0.999 for iQ Supermix). There was no significant difference between the two standard curves according to ANCOVA (P = 0.190), suggesting that either reaction mixture could be used with equal success for qPCR, allowing considerations of convenience and cost to take priority in the choice of this reagent. Because cost is an important consideration when developing a practical detection method for commercial application, it also should be noted that although the manufacturers of both mixes recommend a product volume of 50 μL for a qPCR reaction, we obtained excellent results with only 10 μL, thereby reducing reagent costs by 80% regardless of which product is used.
While the choice of probe set is critical to the experimental design, the qPCR mixture chosen is likely to be less important than the care needed when combining qPCR reaction components.
PC
When duplex qPCR was performed on samples to detect the V. vinifera gene and B. cinerea amplicons simultaneously, once again there was an extremely strong log-linear response between the Ct value and fungal DNA quantity (Figure 3a). Similarly, the PC value was proportional to the quantity of the latter in the sample (Figure 3b).

Results of duplex quantitative polymerase chain reaction (qPCR) assaying various quantities of Botrytis cinerea DNA in the presence of 20 ng of Vitis vinifera DNA extracted from receptacle. (a) Standard curve of B. cinerea amplicon. Threshold cycles (Cts) were plotted against the logarithm of the mass (ng) of a genomic DNA standard with each data point representing the mean of six replications and error bars representing one standard deviation. (b) Pathogen coefficient (PC), the Ct value for the V. vinifera gene divided by that for the B. cinerea amplicon, as a function of the quantity of B. cinerea DNA. Each data point represents the mean of the six replicate from two assays. Error bars represent one standard deviation.
Duplex quantification of plant and pathogen DNA is valuable because it provides an endogenous reference (grape DNA) and allows normalisation for variations caused by differences in DNA extraction among samples, PCR efficiencies and pipetting volumes (Valsesia et al. 2005). Indeed, several recent studies of PCR-based quantitative detection of fungi have included measurements of host plant DNA to provide internal normalisation (Valsesia et al. 2005, Atallah et al. 2007, Brunner et al. 2009, Mideros et al. 2009). Determining PCs might be of particular importance for comparing the amount of B. cinerea DNA from tissues sampled in the vineyard, where there is likely to be even more variation among tissues according to the stage of organ development, crop ripeness, cultivar or location. As these sources of variability could affect DNA quality and qPCR efficiency, such utilisation might also be applicable to studies of disease biology and management under variable field conditions, including evaluation of plant resistance, fungicide efficacy and epidemiological factors.
Quantification of B. cinerea DNA in the receptacle
Consistent with the results earlier for duplex qPCR, addition of B. cinerea conidia to grape receptacle resulted in a strongly linear relationship between the level of B. cinerea amplicons detected and the number of spores in the sample (Figure 4a). The PC increased linearly with increasing numbers of B. cinerea conidia in the sample (Figure 4b).
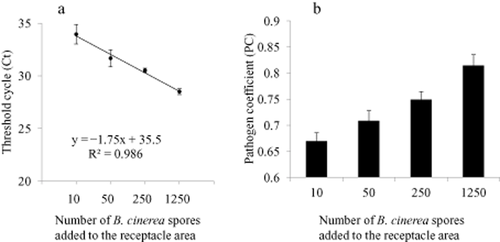
Results of duplex quantitative polymerase chain reaction (qPCR) assaying various numbers of Botrytis cinerea conidia applied to grapevine receptacles. (a) Standard curve of B. cinerea amplicon. Threshold cycles (Cts) were plotted against the logarithm of the number of conidia applied with each data point representing the mean of six replicates and error bars representing one standard deviation. (b) Pathogen coefficient (PC), the Ct value for the Vitis vinifera gene divided by that for the B. cinerea amplicon, as a function of the quantity of B. cinerea conidia applied. Each data point represents the mean of six replications from two assays. Error bars represent one standard deviation.
Application of the technique and future research
At present, qPCR is primarily a research tool for biological studies where differences in pathogen DNA mass are correlated to differences in the extent of pathogen infection and colonisation. The fact that the PC increased linearly with an increasing number of B. cinerea conidia in the receptacle suggests that a positive correlation exists. qPCR could be particularly useful for studying the effects of host or environmental factors that influence pathogen colonisation in time and space before disease becomes visible. Environmental factors include interventions in the vineyard for disease management.
The receptacle area is likely to be an important pathway for infection by B. cinerea (Holz et al. 2003, Keller et al. 2003, Viret et al. 2004, Gindro et al. 2005), and it is the target for crop protection during flowering. Crop protectants are also applied before bunch closure to ensure good spray coverage inside the bunch; however, the effect of these chemicals is not evident until the symptoms of bunch rot develop, usually after veraison. Application of qPCR before and after fungicide treatment might elucidate whether or not a fungicide is acting as a curative treatment, given that infection might have established at flowering or in the period between flowering and fungicide treatment. Results from this study indicate that no barriers to obtaining high-quality qPCR results from the receptacle of pea-sized berries, although collection of pea-sized berries from a range of vineyard environments and cultivars would confirm this finding.
Few tools are available for detection of B. cinerea at the earliest stages of infection, including qPCR, microscopy and culture techniques to promote fungal growth. If qPCR was developed into an application for assessing disease risk, it is unlikely to be used directly by vineyard managers unless it is low cost and easy to implement; while we have improved the per sample cost of qPCR here, its strength currently remains as a tool for biological research. Near infrared spectroscopy (Scott et al. 2010) is an example of a potentially practical and rapid method for B. cinerea quantification; however, methods such as these need to be calibrated, with qPCR providing a potential alternative to visual or cultural detection techniques.
Conclusions
Quantitative PCR was optimised for detection of B. cinerea in pea-sized berries of the ‘Pixie’ mutant of V. vinifera cv. Pinot Meunier using the Bc3 probe set in a duplex assay with endogenous grape DNA. The high copy number of the target B. cinerea gene resulted in a sensitive assay, with a Ct value of <33 when the reaction was initiated with 0.001 ng of the purified B. cinerea DNA. The Bc Taq and UTAS probe sets also amplified single copy genes of B. cinerea reliably and reproducibly when using dilutions of purified B. cinerea DNA, but with less sensitivity. It was demonstrated that the cost of the duplex assay could be reduced significantly by substituting iQ Supermix with the HS Taq reaction mixture in a reaction volume of 10 μL. The DNA extraction method and the probe sets, however, are critical to the success of qPCR. Finally, the use of a PC was demonstrated, which should facilitate a comparison of results obtained from assays that are not run concurrently or for those of different host tissues. The PC increased with an increasing numbers of B. cinerea conidia in grape receptacles supporting the use of qPCR to assess the extent of host tissue colonisation by the pathogen.
Acknowledgements
The authors acknowledge Dr Marc Fuchs, Dr Chris Smart, Judy Burr, Duane Riegel, John Gottula and Dr Kameka Johnson for helpful conversations related to the project and manuscript; Dr Peter Cousins and Dr Gan-Yuan Zhong for providing plant material; and Holly Lange and Nicholas Trask for technical support in qPCR manipulation. Cornell University, USDA-NIFA Viticulture Consortium-East and the New York Wine and Grape Foundation provided support for the project. This work was also supported by Australian grape growers and winemakers through their investment body Grape and Wine Research and Development Corporation (Project No UT0601). Support from the Australian wine sector was matched by the Australian Government.
Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The US Department of Agriculture is an equal opportunity provider and employer.




