Yeast populations associated with grapes during withering and their fate during alcoholic fermentation of high-sugar must
Abstract
Background and Aims
Grape mycobiota may be a determining factor for the population dynamics that develop during alcoholic fermentation for the production of wine. For sweet wine fermentations, high-sugar content grape musts are employed that represent complex microbial ecosystems. The focus of this study, the Passito di Caluso, is a sweet wine produced in the North of Italy from grapes harvested in the fall and subjected to a withering process.
Methods and Results
The withering process was studied by sampling and microbiological analysis, while the alcoholic fermentation was followed by both culture-dependent and culture-independent approaches. During the withering process we observed a succession of three yeast populations associated with grapes. A high degree of species biodiversity was detected the last day of the monitoring period. The dominance of Saccharomyces cerevisiae in the inoculated fermentation was confirmed.
Conclusions
A succession of yeast populations was observed during grape withering; species such as Candida zemplinina, Metschnikowia fructicola and Hanseniaspora uvarum were also detected during alcoholic fermentation. Autochthonous C. zemplinina populations could play an important technological role in sweet wine production.
Significance of the Study
The grape mycobiota during withering was described and its fate during alcoholic fermentation determined by molecular identification methods.
Introduction
Passito wines, a particular typology of sweet wines, are produced from dehydrated grapes with high sugar concentration, and achieved through a drying or withering process, which can be on-vine or off-vine. For the off-plant dehydration, mature grapes are harvested and subjected to drying either directly in the sun or in rooms, under controlled (temperature, relative humidity, air flow) or ambient conditions. Apart from the concentration of sugars, other physicochemical characteristics of the grapes are altered, contributing to the characteristic profile of these sweet wines (Rolle et al. 2011). Further, a contribution in the dehydration process can be provided by the development of Botrytis cinerea, in the form of ‘noble rot’. In this last case, the resulting botrytised wines are enriched with aroma compounds produced by B. cinerea (Magyar 2011). The environmental conditions that develop during the withering process create a particular ecological niche that may apply a selective pressure and determine the microbial ecology that evolves in the grape must. Potential problems associated with sweet wine production may be the sluggish initiation of fermentation (Bisson 1999) and the increased acetic acid production due to Saccharomyces cerevisiae osmotic stress (Erasmus et al. 2003).
The main sources of yeast biodiversity in spontaneous alcoholic fermentation are represented by the grape ecosystem and the resident mycobiota in the winery environment. The importance of the grape mycobiota, especially in the first stages of the alcoholic fermentation, is well documented (Fleet 2003, Prakitchaiwattana et al. 2004, Barata et al. 2012). This aspect connects well with a new trend arising in winemaking, represented by the use of cultures of S. cerevisiae mixed with other non-Saccharomyces species. This strategy has been proven to add complexity to the final product or improve fermentation performance (Bely et al. 2008, Ciani et al. 2010, Rantsiou et al. 2012). The study of the grape mycobiota and of the evolution of yeast populations during subsequent alcoholic fermentation provides valuable information regarding species distribution and persistence. It is the first step in evaluating potential candidates for development of cultures, of autochthonous origin, ideally adapted in specific environments, such as high-sugar content musts.
With this perspective, in this study we focused on the microbiological aspects of the withering process of Erbaluce grapes. This variety is autochthonous to the Piedmont region of Italy and is used for the production of the traditional Passito di Caluso Denominazione d'Origine Controllata e Guarantita (DOCG) wine (Rolle et al. 2012). Withering takes place in the autumn – winter months, in relatively cold conditions. An alcoholic fermentation was followed in order to describe the main yeast populations involved. For this purpose we employed a culture-dependent approach, based on plating and molecular identification of isolates, and a culture-independent approach. Polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) was applied on DNA and RNA extracted directly from fermenting must in order to detect the yeast populations present (DNA) and metabolically active (RNA) during the fermentations.
Materials and methods
Grape withering
Grape clusters of the Erbaluce white cultivar (Vitis vinifera L.) from a vineyard located in Caluso (Piedmont, North-West Italy) were carefully harvested in the 2007 season at a soluble solids content of 25°Brix. About 10 000 kg of grapes were placed in perforated boxes (60 × 40 × 15 cm, 6 kg of grapes in each box) in a single layer. For the natural, off-vine dehydration process, the boxes were placed inside a typical room called fruttaio without control of temperature, relative humidity or air flow (ambient conditions). The fruit was dehydrated for 139 days, from 12 September 2007 to 29 January 2008 (autumn–winter thermohygrometric drying conditions) in accordance with the Erbaluce di Caluso DOCG wine production rules (Rolle et al. 2012). The participating winery in this study undertakes one dehydration process per year on the grapes destined for the production of Passito di Caluso; thereby, one batch was followed in this work.
During this off-vine withering period, grapes were sampled approximately every 15 days (0, 17, 30, 49, 63, 93, 124 and 139 days after harvest). At each sampling time, three lots of about 500 sound berries were sampled randomly, placed in sterile stomacher bags and transported to the laboratory within an hour. The grapes were manually crushed inside the stomacher bag, and the grape juice obtained was subjected to chemical and microbiological analysis.
Must fermentation and sampling
At the beginning of February 2008, the dried grapes were ready for vinification. They were crushed, resulting in a must with a pH value of 3.23 and a titratable acidity of 8.12 g/L expressed as tartaric acid. The must was fermented in triplicate (approximately 1200 L per replicate) by a winery in the Caluso area; a starter culture (Lalvin EC1118, Lallemand, Montreal, Canada) was added and the fermentation lasted 14 days. Before the inoculation of the starter culture, 25 mg/L SO2 was added. The fermentation was conducted at a controlled temperature of 23 ± 1°C and was sampled at 1, 3, 7 and 14 days. At each sampling point 50 mL of the must and fermenting must was collected in sterile screw cap tubes and stored at 4°C during transportation to the laboratory. Microbiological analysis was carried out as described later. In addition, at each sampling point, aliquots (1 mL each) of the samples were collected for chemical analysis and for the extraction of DNA and RNA. The aliquot destined for chemical analysis was filtered through a 0.2-μm filter and stored at −20°C until analysis. The aliquots destined for DNA and RNA extraction were centrifuged for 5 min at 13 400 rpm, the supernatant was removed and for the RNA aliquot, 0.2 mL of RNAlater (Ambion, Milan, Italy) was added. Pellets prepared in this way were stored at −20°C.
Microbiological analysis
For microbiological analysis, serial dilutions in Ringer's solution were prepared and plated on the Wallerstein Laboratory Nutrient medium (for grapes during withering and must during fermentation), a differential medium on which yeast colonies can putatively be identified based on their topomorphology (Pallmann et al. 2001, Urso et al. 2008), and on the Lysine medium (for must during fermentation) to count non-Saccharomyces (Oxoid, Milan, Italy). Plates were incubated at 30°C for 5 days and subsequently counted.
Chemical analysis
Glucose, fructose, malic acid, glycerol, ethanol, and acetic acid of grape juices and fermenting musts were quantified by means of a high-performance liquid chromatograph (HPLC) (Thermo Electron Corp., Waltham, MA, USA) equipped with a UV detector (UV100), set to 210 nm, and a refractive index detector (RI-150). The analyses were run isocratically at 0.8 mL/min and 65°C on a cation-exchange column (300 mm by 7.8 mm inner diameter; Aminex HPX-87H) fitted with a Cation H+ Microguard cartridge (Bio-Rad Laboratories, Hercules, CA, USA), using 0.0026 N H2SO4 as the mobile phase (Giordano et al. 2009).
Molecular identification of isolates
At each sampling point during the grape withering process and the fermentation, yeast colonies (at least 10) were randomly selected from the WLN medium, isolated and stored at −80°C in yeast-peptone-dextrose broth (YPD, 2% (wt/vol) glucose, 2% (wt/vol) peptone and 1% (wt/vol) yeast extract, all from Oxoid, Milan, Italy), with glycerol (30% final concentration). For the molecular identification, isolates were grown in YPD broth and their DNA was extracted, amplified with primers NL1GC/LS2 and subjected to denaturing gradient gel electrophoresis (DGGE) (Cocolin et al. 2000) (as described later). Isolates were grouped according to their DGGE profile, and representatives of each group were amplified with primers NL1/NL4 (Kurtzman and Robnett 1997) to obtain a polymerase chain reaction (PCR) product, which was sequenced by a commercial facility (Eurofins, Edersberg, Germany). Identification of each group was based on a BLAST search (Altschul et al. 1990) of the sequence obtained on the National Center for Biotechnology Information. Both primer pairs targeted the D1-D2 loop of the 26S rRNA gene.
Nucleic acids extraction from must
The protocols described in Mills et al. (2002) were followed for the extraction of DNA and RNA from must; DNA was quantified with a Nanodrop ND-1000 spectrophotometer (Celbio, Milan, Italy) and standardised to 100 ng/μL. In order to eliminate DNA traces from the preparation, RNA was resuspended in 50 μL of water containing the Turbo DNase (Ambion, Milan, Italy). Complete DNA digestion was confirmed by using 1 μL in PCR, and if a product was obtained, the treatment was prolonged until negative PCR reaction was obtained from all RNA samples.
PCR and reverse transcription (RT)-PCR amplification
Amplification of the DNA (extracted from pure cultures or directly from the must) was achieved with primers NL1 (5′-GCC ATA TCA ATA AGC GGA GGA AAA G-3′) and LS2 (5′-ATT CCC AAA CAA CTC GAC TC-3′) (Cocolin et al. 2000). A GC-clamp (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-3′) was attached to the forward NL1 primer when the PCR product was destined for DGGE analysis (Sheffield et al. 1989). PCR was performed in a final volume of 25 μL containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1.25 U of Taq Polymerase (Applied Biosystems, Milan, Italy) and 0.2 μM of each primer. A total of 100 ng of DNA was added in the reaction. Amplifications were carried out in a PTC-220 DNA Engine Dyad MJ Research thermalcycler (Celbio). The amplification cycle was denaturation at 95°C for 1 min, annealing at 42°C for 1 min and extension at 72°C for 1 min, and the cycle was repeated 35 times. The cycle was preceded by an initial denaturation at 95°C for 5 min and followed by a final extension at 72°C for 7 min.
Reverse transcription was performed with the Moloney murine leukemia virus reverse transcriptase of Promega (Milan, Italy). One microgram of RNA was mixed with 100 μM of primer LS2 in a final volume of 10 μL. The mix was denatured at 75°C for 5 min, immediately put on ice and then the RT reaction mix was added. The RT was performed in the 25 μL total volume containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 2 mM dNTPs, 4 μM primer, 200 units of M-MLV and 0.48–0.96 units of RNasin ribonuclease inhibitor. The mix was incubated at 42°C for 1 h, and it was followed by a regular PCR reaction, as described earlier (addition of 1 μL of RT reaction into 25 μL of PCR reaction).
Denaturing gradient gel electrophoresis
After agarose gel electrophoresis (2% in 1.25 X Tris-Acetate-EDTA), PCR products were analysed by DGGE, using the D-Code universal mutation detection system (Bio-Rad Laboratories), with a 0.8-mm thick polyacrylamide gel (8% (wt/vol) acrylamide-bisacrylamide (37.5 : 1)). A 30–50% denaturing gradient (100% corresponds to 7 mol urea and 40% (wt/vol) formamide), increasing in the direction of the electrophoretic run, was used. The run was undertaken at 60°C using 130 V for 270 min. Gels were stained for 20 min in 1.25 X Tris-acetate-EDTA containing 1 X SYBR Green (Sigma, Milan, Italy). They were visualised under UV light, digitally captured and analysed with the UVIpro Platinum 1.1 Gel Software (Eppendorf, Milan, Italy) for the recognition of the bands present.
Results
Microbial counts and yeast biodiversity during grape withering
Yeast and mould counts on grapes during withering (Table 1) followed a similar trend. They decreased to their lowest level on day 63, followed by an approximately 2-log10 cfu/mL increase at day 93 with no further change until the end of the monitoring period. Yeast counts fluctuated between 3.63 log10 cfu/mL (lowest level recorded after 49 days of withering) and 5.71 log10 cfu/mL (highest count reached at day 93). Overall, yeast counts at the beginning and end of the process remained constant and in the order of 4 log10 cfu/mL. The range for moulds was between 2.76 log10 cfu/mL (lowest count, day 30) and 4.72 log10 cfu/mL (highest count, recorded at day 0). The count at the end of the period was 4.37 log10 cfu/mL for yeasts and 3.18 log10 cfu/mL for moulds.
| Day 0 (harvest) | Day 17 | Day 30 | Day 49 | Day 63 | Day 93 | Day 124 | Day 139 | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Log10 yeast cfu/mL ± SDa | 4.68 ± 0.62 | 4.75 ± 1.52 | 4.31 ± 1.73 | 3.63 ± 0.86 | 3.81 ± 1.02 | 5.71 ± 1.56 | 4.36 ± 0.82 | 4.37 ± 0.56 | |
| Log10 mould cfu/mL ± SDa | 4.72 ± 0.15 | 4.10 ± 0.62 | 2.51 ± 0.06 | 3.15 ± 0.6 | 2.76 ± 0.18 | 4.68 ± 0.32 | 3.67 ± 0.51 | 3.18 ± 0.19 | |
| Hanseniaspora uvarum | 8 (66.7) | 6 (42.8) | 7 (46.7) | 4 (44.4) | 5 (26.3) | 4 (28.6) | 2 (5.1) | 36 (27.1) | |
| Aureobasidium pullulans | 1 (8.3) | 3 (20) | 4 (21) | 2 (18.2) | 10 (71.4) | 20 (51.3) | 40 (30.1) | ||
| Candida zemplinina | 2 (14.3) | 5 (20) | 4 (21) | 5 (45.5) | 6 (15.4) | 22 (16.5) | |||
| Rhodotorula nothofagi | 1 (8.3) | 2 (14.3) | 4 (21) | 7 (5.3) | |||||
| Rhodotorula glutinis | 1 (9.1) | 1 (0.7) | |||||||
| Candida californica | 2 (18.2) | 2 (1.5) | |||||||
| Isatchenkia terricola | 1 (8.3) | 1 (0.7) | |||||||
| Metschnikowia fructicola | 1 (8.3) | 4 (28.6) | 5 (55.6) | 2 (10.5) | 1 (9.1) | 3 (7.7) | 16 (12) | ||
| Pichia anomala | 1 (2.5) | 1 (0.7) | |||||||
| Lachancea thermotolerans | 2 (5.1) | 2 (1.5) | |||||||
| Saccharomyces cerevisiae | 4 (10.2) | 4 (3) | |||||||
| Candida ishiwadae | 1 (2.5) | 1 (0.7) | |||||||
| Total isolates for each sampling point | 12 | 14 | 15 | 9 | 19 | 11 | 14 | 39 | 133 |
- a Counts presented are the mean ± standard deviation of triplicate samples at each time point.
At each sampling point, colonies on the WLN medium were randomly picked, isolated and identified to the species level by PCR-DGGE grouping and sequencing of partial 26S rRNA encoding gene (Table 1). A total of 133 isolates, belonging to 12 species, was identified. The species most represented were Aureobasidium pullulans, with 40 isolates, followed by Hanseniaspora uvarum, with 36 isolates, and Candida zemplinina, with 22 isolates. These three species, together with Metschnikowia fructicola, were also constantly present on the grapes. Hanseniaspora uvarum was isolated in seven out of eight sampling points, while A. pullulans and M. fructicola were isolated in six out of eight time points, and C. zemplinina in five out of eight. All four species were present both on the first and the last day of sampling. Hanseniaspora uvarum was predominant during the first period of withering, up to day 49, and constituted almost 50% of the isolates, while A. pullulans prevailed at the end of the period, reaching 71.4 and 51.3% of the isolates at 124 and 139 days, respectively. Rhodotorula nothofagi, Rhodotorula glutinis, Candida californica and Issatchenkia terricola constituted minor populations during the monitoring period because they were isolated at a low proportion and unsystematically. Finally, four species, Pichia anomala, Lachancea thermotolerans, S. cerevisiae and Candida ishiwadae, were isolated only on the last day of the monitoring period (day 139).
Microbial counts and yeast biodiversity during fermentation
The total yeast counts and the non-Saccharomyces counts during the fermentation are presented in Table 2. A yeast count of about 5 log10 cfu/mL was recorded for the grape must used for the fermentation. On day 1 a 2-log10 increase was observed and the counts stabilised to around 8 log10 cfu/mL for the remainder of the period (days 3–14). The initial non-Saccharomyces count (in the grape must) was of the order of 4 log10 cfu/mL, and it remained stable throughout the fermentation. Similarly to that described for grapes during drying, yeasts were randomly isolated and identified during the fermentation. In the grape must, six species were identified, including S. cerevisiae. Except for Pichia kluyveri, which was only detected in the grape must, all other species were common between the grape sample at day 139 and the grape must sample. In the fermentation, S. cerevisiae dominated the fermentation and represented 49% (37 isolates out of 75 in total) of the isolates. It was present from day 1 up until the last day of the fermentation. Non-Saccharomyces yeasts were present up until day 7 of the fermentation and belonged to P. kluyveri, C. zemplinina, M. fructicola, H. uvarum, A. pullulans, P. anomala, C. ishiwadae and L. thermotolerans, in order of abundance.
| Grape must | Days after start of fermentation | Total isolates | ||||
|---|---|---|---|---|---|---|
| 1 | 3 | 7 | 14 | |||
| Log10 yeast cfu/mL ± SDa | 4.99 ± 0.27 | 7.17 ± 0.33 | 8.30 ± 0.15 | 8.96 ± 0.05 | 8.54 ± 0.29 | |
| Log10 non-Saccharomyces yeast cfu/mL ± SDa | 4.39 ± 0.11 | 5.43 ± 0.6 | 5.78 ± 0.08 | 5.22 ± 0.04 | 4.12 ± 0.33 | |
| Pichia anomala | 1 (4.5) | 3 (10.7) | 4 (5.3) | |||
| Candida zemplinina | 4 (13.3) | 1 (4.5) | 4 (14.3) | 5 (41.7) | 10 (13.3) | |
| Metschnikowia fruticola | 3 (10) | 1 (4.5) | 3 (10.7) | 4 (5.3) | ||
| Lachancea thermotolerans | 1 (3.6) | 1 (1.3%) | ||||
| Saccharomyces cerevisiae | 3 (10) | 7 (31.8) | 11 (39.3) | 6 (50) | 13 (100) | 37 (49.3) |
| Hanseniaspora uvarum | 7 (23.3) | 1 (3.6) | 1 (8.3) | 2 (2.7) | ||
| Aureobasidium pullulans | 2 (9.1) | 2 (7.1) | 4 (5.3) | |||
| Candida ishiwadae | 6 (20) | 1 (3.6) | 1 (1.3) | |||
| Pichia kluyveri | 7 (23.3) | 10 (45.5) | 2 (7.1) | 12 (16) | ||
| Total of isolates for each sampling point | 30 | 22 | 28 | 12 | 13 | 75 |
- a Counts are the mean ± standard deviation of the three independent fermentations followed. Yeast counts were determined on Wallerstein Laboratory Nutrient medium plates, while non-Saccharomyces on the Lysine medium. SD, standard deviation.
Population dynamics and evolution during alcoholic fermentation by PCR-DGGE
Total DNA and RNA were extracted from the grape sample at the end of the withering period (day 139), from the grape must used for fermentation as well as during the alcoholic fermentation (days 1, 3, 7 and 14). The nucleic acids were used as templates for PCR-DGGE, using primers NL1-LS2, in order to profile the yeast populations at each sampling point. The DGGE profiles of the samples of grapes and grape must at the DNA and RNA level are shown in Figure 1. No differences were observed between the three replicates of the fermentation; only one is presented. The profiles obtained from the grapes and grape must were similar. At the DNA level, the profile was characterised by the presence of B. cinerea and another fungal species, which was not possible to be identified more precisely. The other two bands that were visible in these samples belonged to C. zemplinina. Also present at the RNA level was C. zemplinina, together with A. pullulans, in grapes and grape must. During fermentation, from day 1 onwards, the profiles presented bands appertaining to S. cerevisiae only, at both DNA and RNA levels (data not shown).
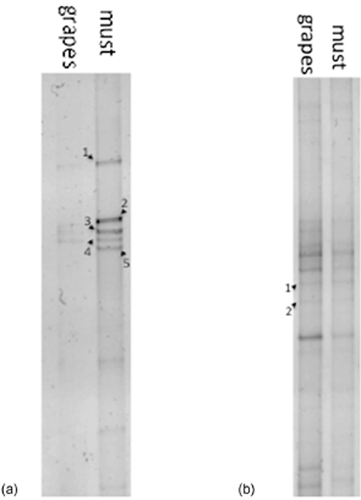
DNA (a) and RNA (b) denaturing gradient gel electrophoresis (DGGE) profiles of samples from grapes at the end of the withering period (day 139) and grape must. Bands marked with a number were excised and sequenced (as described in the materials and methods section). Band identification for panel a: 1 and 3, Botrytis cinerea; 2 and 5, Candida zemplinina; 4, Fungal sp. Band identification for panel b: 1, Candida zemplinina; 2, Aureobasidium pullulans.
Chemical analysis
The results of the chemical analysis of the process of grape withering and of the alcoholic fermentation are shown in Figure 2 and Table 3, respectively.
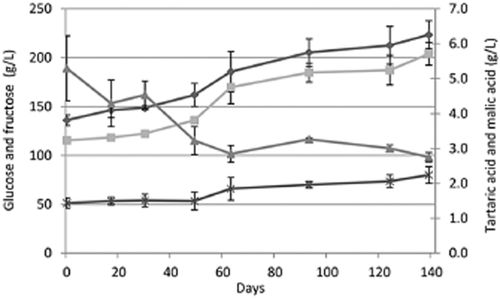
The concentration of glucose ( ), fructose (
), fructose ( ), malic acid (
), malic acid ( ) and tartaric acid (
) and tartaric acid ( ) during grape withering. For each sampling point the mean and standard deviation of the chemical analysis performed on triplicate samples are presented.
) during grape withering. For each sampling point the mean and standard deviation of the chemical analysis performed on triplicate samples are presented.
| Grape must | 1 | 3 | 7 | 14 | |
|---|---|---|---|---|---|
| Glucose (g/L) | 221 ± 2 | 206 ± 1 | 174 ± 5 | 117 ± 1 | 76 ± 1 |
| Fructose (g/L) | 207 ± 6 | 196 ± 1 | 181 ± 6 | 155 ± 2 | 132 ± 1 |
| Ethanol (mL/L) | 3.0 ± 0.8 | 11.5 ± 0.1 | 32.1 ± 0.5 | 86.9 ± 2.1 | 133.1 ± 0.6 |
| Glycerol (g/L) | 3.9 ± 0.1 | 4.0 ± 0.1 | 7.8 ± 0.1 | 12.7 ± 0.2 | 14.2 ± 0.1 |
| Acetic acid (g/L) | 0.06 ± 0.03 | 0.06 ± 0.01 | 0.38 ± 0.01 | 0.75 ± 0.02 | 0.88 ± 0.01 |
| Malic acid (g/L) | 3.5 ± 0.1 | 3.3 ± 0.1 | 3.3 ± 0.1 | 2.6 ± 0.1 | 2.5 ± 0.1 |
A progressive increase of sugar content, proportionally to weight loss, was observed during the grape dehydration process. In accordance with a previous study (Rolle et al. 2011), a significant decrease of tartaric acid was noticed, in particular during the first 60 days of withering. B. cinerea Pers. has the property of degrading the tartaric acid (Ribéreau-Gayon et al. 2000). The ambient conditions of this withering process are favourable for the development of Botrytis at the stage of noble rot (Rolle et al. 2012). In fact, the glycerol concentration in the grape must, a chemical marker of Botrytis infection (Ravji et al. 1988), was 3.9 g/L (Table 3).
The final must used for the fermentations was characterised by a high sugar content (428 g/L of sugars). Glucose and fructose were consumed in parallel, although at different speed. During fermentation, glucose was consumed at a higher rate than that of fructose. From a starting value of about 221 g/L, it reached a final value of about 76 g/L. Fructose, in contrast, was reduced from about 207 to 132 g/L. The ethanol content at the end of fermentation was on average 133 mL/L (ethanol produced/consumed sugar yield 0.466 ± 0.004 (g/g)). During fermentation about 11 g/L of glycerol was produced. The malic content of the final wine was 1 g/L less than that of the grape must.
Discussion
Numerous authors have reported on the importance of grape yeast ecology and its influence on wine quality. Grapes are the main source of yeasts that inoculate must and initiate the alcoholic fermentation. Non-inoculated alcoholic fermentations rely on yeast biota, indigenous to the must, while in inoculated fermentations, initial stages of the process are characterised by the coexistence of wild, usually non-Saccharomyces yeasts and those of the starter culture. The purpose of this work was twofold. First, we investigated the yeast ecology on mature grapes of the Erbaluce cultivar during off-vine withering. Then, we focused on monitoring the yeast dynamics and the principal chemical parameters of an inoculated fermentation of the high sugar must. In this study, particular attention was given to culture-independent methods, able to study DNA and RNA extracted directly from the matrix without isolation of the yeasts. Studying the DNA, it is possible to define the number and identity of the microbial species present in a specific sample, thereby giving a view of the microbial diversity and ecology. In contrast, RNA gives insight into the metabolically active portion of the populations present. This is relevant for food fermentations, including alcoholic fermentation for wine production, where it is necessary to study the species that are responsible for the transformation (Cocolin et al. 2011). It should be emphasised that the detection limit for DGGE is of the order of 103 cfu/mL (Cocolin et al. 2000). As a consequence, microbial groups that are present and active, but whose population is lower than 103 cfu/mL, will not be taken into consideration. The determination of the detection limit in DGGE analysis is not always simple to perform because it depends on the different affinity that the primers have towards the microbial species present in one ecosystem; thereby, this limit may change according to the specific group of microorganisms being studied.
Three species successively dominated the grape mycobiota. Hanseniaspora uvarum prevailed for about the first half of the grape withering process, and although it was present until the last day of sampling, its frequency of isolation decreased in the second half of the process. Concomitantly, C. zemplinina and A. pullulans, occasionally isolated at the beginning, became the major component of the mycobiota towards the end of the process. Among the first reports on the dominance and persistence of A. pullulans on wine grapes is that of Prakitchaiwattana et al. (2004), in a pioneer study of grape ecology by a combination of culture-dependent and culture-independent approaches. Subsequently, other authors have also reported on its presence (Renouf et al. 2005, Nisiotou and Nychas 2007). Aureobasidium pullulans is known to possess antagonistic properties towards other yeasts and fungi, and it can be speculated that it may influence the overall grape ecology (Castoria et al. 2001, Prakitchaiwattana et al. 2004). A signal, corresponding to A. pullulans, was also detected on the grapes sampled on the last day of withering, at the RNA level.
Since its description in 2003 (Sipiczki 2003), C. zemplinina has been frequently isolated or detected in alcoholic fermentations (Urso et al. 2008, Tofalo et al. 2009), while several strains, isolated from grapes and previously identified as C. stellata, are now reclassified as C. zemplinina (Csoma and Sipiczki 2008). The psychrotolerant and osmotolerant properties that C. zemplinina strains posses can be advantageously exploited in botrytised-sweet wines production, characterised by a high sugar concentration and a low fermentation temperature (Sipiczki 2004). Furthermore, the possibility to combine C. zemplinina with S. cerevisiae, in order to alleviate the osmotic stress imposed on S. cerevisiae in high-sugar musts and in this way reduce acetic acid production, was recently investigated (Rantsiou et al. 2012). For these reasons, the consistent presence of C. zemplinina during the withering process of the grapes studied here is important. The grapes are carriers of this microbial species and may have an important technological impact for the alcoholic fermentation and the production of Passito di Caluso sweet wine. This aspect was confirmed during the monitoring of the fermentation, where C. zemplinina strains were isolated up to day 7.
An interesting finding of this study that regarded the grape ecology was the detection at different time points of the withering process, of a recently described species, M. fructicola. Metschnikowia fructicola is a close relative of M. pulcherrima, a species that is associated with grapes (Barata et al. 2012), may persist in grape must during the first days of fermentation and has been described to possess antifungal activity (De Curtis et al. 1996). Recently, Metschnikowia strains isolated from botrytised grapes were tested and resulted to be antagonistic towards fungal and bacterial growth (Sipiczki 2006). This finding suggests that the presence of the Metschnikowia species, specifically M. pulcherrima and M. fructicola, may play a protective role against the growth of filamentous fungi on the grapes during withering (Kurtzman and Droby 2001).
The largest biodiversity in terms of species detected was observed on the grapes on the last day of withering. Then, once the grapes were crushed, there was a decrease in the number of species detected, from eight in the grapes to six in the must. Candida zemplinina, H. uvarum, M. fructicola, S. cerevisiae and C. ishiwadae were species common to the grapes and the grape must. Differences were observed also in the population dynamics, as determined by the culture-dependent method, during the course of the fermentation. Saccharomyces cerevisiae was the dominant population, representing 49% of the isolates, and was isolated from the first to the last day of fermentation. It is surprising that although the must was inoculated with a starter culture, several other species could be detected up to day 7 of fermentation (when the alcohol content was 87 mL/L), indicating a persistence of yeast populations other than the starter, as reported previously (Urso et al. 2008, Rantsiou et al. 2012). Among them, C. zemplinina and H. uvarum were constant. It should be noted that by culture-independent analysis, a signal corresponding only to S. cerevisiae was detected as reported earlier (data not shown).
Conclusions
To our knowledge, this is the first time that the mycobiota has been studied throughout the grape withering process and that its fate has been followed during alcoholic fermentation. Although a single batch of grapes was followed during the withering process, because all grapes used for the Passito di Caluso wine were treated at the same time, it is not possible to assess if the ecology found would be confirmed in additional studies; however, it is important to underline the importance of the results obtained in terms of the contribution of yeasts developed on the grapes to the fermentation. During the withering process a succession of three populations associated with grapes was observed, while a high degree of species biodiversity was detected at the end of the monitoring period. The significance of C. zemplinina in sweet wine fermentations was confirmed; it was a numerically important part of the yeast mycobiota during fermentation, and its provenance was the grapes. Generally the results between culture-dependent and culture-independent approaches used in this study compared well.
Acknowledgement
The authors wish to thank the technical staff of the winery that provided the samples of grapes during withering and of the fermenting must for this study.




