An identical-by-descent novel splice-donor variant in PRUNE1 causes a neurodevelopmental syndrome with prominent dystonia in two consanguineous Sudanese families
Authors Mahmoud Koko, Ashraf Yahia, and Liena E. Elsayed contributed equally.
Abstract
PRUNE1 is linked to a wide range of neurodevelopmental and neurodegenerative phenotypes. Multiple pathogenic missense and stop-gain PRUNE1 variants were identified in its DHH and DHHA2 phosphodiesterase domains. Conversely, a single splice alteration was previously reported. We investigated five patients from two unrelated consanguineous Sudanese families with an inherited severe neurodevelopmental disorder using whole-exome sequencing coupled with homozygosity mapping, segregation, and haplotype analysis. We identified a founder haplotype transmitting a homozygous canonical splice-donor variant (NM_021222.3:c.132+2T > C) in intron 2 of PRUNE1 segregated with the phenotype in all the patients. This splice variant possibly results in an in-frame deletion in the DHH domain or premature truncation of the protein. The phenotypes of the affected individuals showed phenotypic similarities characterized by remarkable pyramidal dysfunction and prominent extrapyramidal features (severe dystonia and bradykinesia). In conclusion, we identified a novel founder variant in PRUNE1 and corroborated abnormal splicing events as a disease mechanism in PRUNE1-related disorders. Given the phenotypes’ consistency coupled with the founder effect, canonical and cryptic PRUNE1 splice-site variants should be carefully evaluated in patients presenting with prominent dystonia and pyramidal dysfunction.
1 INTRODUCTION
PRUNE1-related disorders are a group of neurodevelopmental and neurodegenerative diseases caused by homozygous and compound heterozygous alterations in PRUNE1. This gene, located on chromosome 1q21.3, is highly expressed in the human fetal brain, where it participates in neuronal cell proliferation, migration, and differentiation (Kobayashi et al., 2006; Reymond et al., 1999). The canonical transcript (NM_021222) consists of eight exons coding 453 amino-acids. The PRUNE1 protein (prune exopolyphosphatase 1), an enzyme that contains conserved Aspartate–Histidine–Histidine (DHH) and DHH-associated motif 2 (DHHA2) domains hydrolyzes a broad spectrum of substrates. It also interacts with key proteins involved in signaling and cytoskeletal organization, where its role in microtubule organization is directly related to its pathogenicity in neurodevelopmental disorders (Zollo et al., 2017) as well as cancer metastasis (D'Angelo et al., 2004). Although missense variants may increase or decrease the enzymatic activity of PRUNE1, they consistently impair microtubule polymerization (Zollo et al., 2017).
Previously reported pathogenic PRUNE1 missense variants spread over the two phosphodiesterase domains, DHH and DHH2. Recessively inherited missense variants located in the DHH domain provided the first evidence for the involvement of PRUNE1 in a neurodevelopmental disorder characterized by microcephaly, developmental delay or regression, seizures, cortical and cerebellar atrophy, and thin or hypoplastic corpus callosum (Karaca et al., 2015). Afterward, multiple patients harboring missense variants in both the DHH and DHHA2 domains and presenting similar phenotypes were identified (Costain et al., 2017; Zollo et al., 2017). Later work revealed that the microcephaly, frequently reported in PRUNE1-related neurodevelopmental syndrome, has a progressive nature (Baple et al., 2017). However, normal head circumference and macrocephaly were also detected in patients with PRUNE1-related disorder (Alhaddad et al., 2018; Imagawa et al., 2018; Milone et al., 2021). A spinal muscular atrophy-like presentation was reported in two families of Italian and Turkish origins carrying a recurrent homozygous NM_021222.3:c.316G > A (p.(D106N)) variant (Iacomino et al., 2018; Karaca et al., 2015; Okur et al., 2019). Recently, new magnetic resonance findings were suggested to be of potential diagnostic value (Fujii et al., 2020). At least 12 PRUNE1 variants were identified in around 30 families from different geographical origins. About half of these variants were missense alterations clustered around the DHH domain. At least four truncating variants were reported (Fig. 1). The detection of truncating and frame-shift variants as well as a large deletion supported the importance of loss-of-function as a disease mechanism in PRUNE1-linked disorders. A recent study elucidated the detrimental roles of PRUNE1 hypomorphic variant alleles in the pathogenesis of PRUNE1-linked disorders, underscoring loss-of-function as the pathogenic mechanism of these diseases and the potential key role of PRUNE1 exopolyphoshatase activity in neurodevelopment (Nistala et al., 2021).
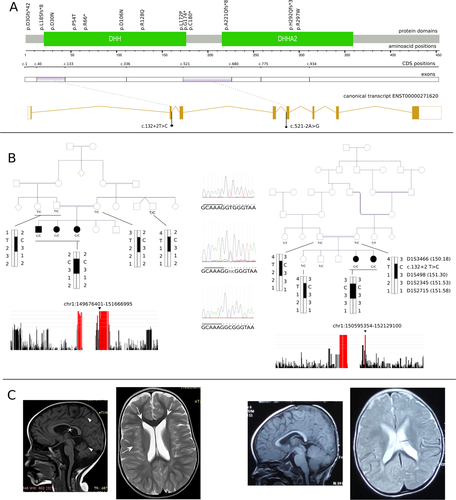
To our knowledge, there has been a single report of a pathogenic PRUNE1 splice-site variant to date. A homozygous canonical splice-acceptor variant in intron 4 (NM_021222.3:c.521-2A > G) was identified first in an Oji-Cree patient with severe global developmental delay, cortical blindness, and infantile spasms (Costain et al., 2017). The same variant was reported in a homozygous state in multiple families from Canada of Cree origin, where it caused a neurodegenerative disease affecting both the peripheral and central nervous systems (Hartley et al., 2019). Here, we report two families from Sudan in which five affected individuals inherited a novel homozygous founder haplotype, highlighting the pathogenicity of in-frame exon skipping and the relevance of splice-site alterations in PRUNE1-associated syndrome.
2 MATERIALS AND METHODS
2.1 Ethics Approval and Consent to Participate
Informed written consent for participation was obtained from participants (or parents of children). Ethical approval was obtained from the ethical committee of the Faculty of Medicine, University of Khartoum. All procedures in this study were performed following the ethical standards of the ethical committee of the Faculty of Medicine, University of Khartoum, Sudan, and with the 1975 Helsinki declaration and its later amendments.
2.2 Phenotyping and Sampling
The patients were evaluated at the Pediatrics Neurology Clinic, Soba University Hospital (Khartoum, Sudan). Saliva samples were collected from the participants using Oragene OG500 kits (DNA Genoteck, Canada). DNA was extracted according to the PrepIT L2P protocol provided by the manufacturer.
2.3 Genetic Analysis
2.3.1 Exome sequencing and array Genotyping
Exome sequencing in family 1
Exome sequencing was performed for two affected individuals at the Brain Institute - ICM (Paris). Enrichment was done using SeqCapEZ v3 (Roche, Basel, Switzerland). Paired-end whole-exome sequencing was performed using the NextSeq500 platform (Illumina, San Diego, CA, USA). Read alignment to the human genome GRCh37 was done using bwa v0.7.4, and variant calling was done using GATK v3.3 (DePristo et al., 2011). Homozygosity mapping was performed with Homozygosity Mapper (Seelow et al., 2009) using exome data from the two patients.
Exome sequencing and array genotyping in family 2
Exome sequencing was performed for a single patient at the Cologne Center for Genomics (CCG) (Cologne, Germany). Enrichment was done using Sure Select XT Human All Exon V6 kits (Agilent, Santa Clara, CA, USA). Paired-end sequencing was done on an Illumina HiSeq4000 platform (Illumina, San Diego, CA, USA). The reads were aligned to the human genome GRCh37 using bwa v0.6.2, and the variants were called using GATK UnifiedGenotyper v1.6. The alignment, variant calling, and annotation were done using the Varbank platform (https://varbank.ccg.uni-koeln.de/). Genome-wide microarray genotyping was performed for seven individuals (two patients, parents, one sibling, and two relatives) at the CCG using the Affymetrix Precision Medicine Research Array (Thermo Fisher Scientific, Waltham, MA, USA). Genotype calling was performed according to the manufacturer`s protocol using Axiom Power Tools (Thermo Fisher Scientific). Homozygosity mapping was performed with Homozygosity mapper (Seelow et al., 2009) using genome-wide array calls from two affected individuals, their parents, and additional controls.
2.3.2 Variant prioritization
We filtered low-quality calls (depth < 20, mapping quality < 30, overlapping known repeat sites). Nonsense, frame-shift, and canonical splice-site alterations, as well as in-frame indels, and missense variants with a CADD score > 20 were prioritized (Rentzsch et al., 2019). We filtered for variants with gnomAD minor allele frequency < 0.1% in the total gnomAD v2 exomes and less than two homozygous calls (Karczewski et al., 2020). We prioritized variations in known neurodevelopmental disorders genes that are in a homozygous run in all the patients.
2.3.3 Validation and segregation
Sanger sequencing was performed on an ABI3730 sequencer (Applied Biosystems) at GATC Biotech – Eurofins Genomics (Cologne, Germany).
2.3.4 Haplotype analysis
Multiple polymorphic short tandem repeat (STR) markers spanning the region of homozygosity (ROH) around PRUNE1 (D1S3466, D1S498, D1S2345, D1S2715) were genotyped in multiple members from both families. The microsatellites were polymerase chain reaction (PCR) amplified using HEX-labeled primers, and the size was estimated using a ROX500 size standard (Eurofins Genomics). The haplotypes were then reconstructed manually.
3 RESULTS
3.1 Clinical Presentation
We examined five patients from two unrelated consanguineous Sudanese families (three siblings from the first family and two sisters from the second family) showing global neurodevelopmental delay, pyramidal, and extrapyramidal signs and symptoms. There was no family history of similar conditions in both families. The presentation was characterized by a perinatal onset of a profound developmental disorder (prenatal in one patient, manifesting with decreased fetal movements, and early neonatal in the others). Global developmental delay, pyramidal (bilateral upper and lower limb spasticity, hyperreflexia, and bilateral extensor plantar response), and extrapyramidal signs (severe dystonia and bradykinesia) were the main features of the clinical phenotype (Table 1). Contractures and musculoskeletal deformities were seen in all patients with different degrees of severity (pes cavus, scoliosis, flexion deformities); however, no cerebellar signs were detected. Optic atrophy was seen in all three patients from the first family, while focal seizures were reported in one patient from the second family. Head circumference was normal in one family and not documented in the other. Neurodegenerative features, including mild cortical, subcortical and cerebellar atrophy, and/or thin corpus callosum, were found in some patients’ brain imaging studies (Fig. 1). Periventricular subcortical white matter hyperintensities were also detected in one affected individual. The details of the clinical presentation are presented in Table 1.
| Feature (abnormalities’ frequency in the literature)* | First family | Second family | |||
|---|---|---|---|---|---|
| Sex | M | F | F | F | F |
| Age at onset | Prenatal (end of the third trimester) | Birth | Birth | Birth | Birth |
| Age at examination | 7 years | 4 years | 2 years | 11 years | 5 years |
| Prenatal development and birth (rare) | Decreased fetal movements and normal delivery | Normal fetal movements and normal delivery | Normal fetal movements and normal delivery | Normal fetal movements and normal delivery | Normal fetal movements and normal delivery |
| Development (very common) | Delayed psychomotor development; never spoke | Delayed psychomotor development; never spoke | Delayed psychomotor development; never spoke | Delayed head support, delayed motor development; never spoke | Delayed motor development; never spoke |
| Pyramidal signs (very common) | Spasticity; diffused hyperreflexia in the upper and lower limbs; positive Hoffmann's sign; up-going plantar response | Spasticity; diffused hyperreflexia in the upper and lower limbs; positive Hoffmann's sign; up-going plantar response | Spasticity; diffused hyperreflexia in the upper and lower limbs; positive Hoffmann's sign; up-going plantar response | Spasticity (stiffness noted at 2 years); diffused hyperreflexia in the upper limbs; positive Hoffmann sign; hyperreflexia in the lower limbs and up-going plantar response; contractures | Spasticity (stiffness noted at 2 years); hyperreflexia (jaw jerk, upper and lower limbs); positive Hoffmann's sign and up-going plantar response; contractures |
| Extrapyramidal signs (ultra-rare) | Severe dystonia and bradykinesia | Severe dystonia and bradykinesia | Severe dystonia and bradykinesia | Severe dystonia | Severe dystonia |
| Musculoskeletal (common) | Bilateral pes cavus and mild scoliosis | Bilateral pes cavus and mild scoliosis | Bilateral pes cavus and mild scoliosis | Severe upper and lower limb contractures; femur fracture | Severe upper and lower limb contractures |
| Head circumference (rare) | Not available | Not available | Not available | 50.5 cm (>3rd percentile) at the age of 11 years | 48 cm (>3rd percentile) at the age of 5 years |
| Dysmorohic features (common) | Absent | Absent | Absent | Absent | Absent |
| Seizures (common) | Absent | Absent | Absent | Focal epilepsy (onset at 5 years); left-sided intermittent runs of spikes on EEG; not on antiepileptic medications | Two attacks of febrile seizures (1 and 2 years of age); normal EEG |
| Eye movement and fundoscopy (rare) | Normal saccadic and pursuit eye movements; optic atrophy | Normal saccadic and pursuit eye movements; optic atrophy | Normal saccadic and pursuit eye movements; optic atrophy | Normal saccadic and pursuit eye movements; normal fundi | Normal saccadic and pursuit eye movements; normal fundi |
| Cerebellar signs (not reported previously) | Absent | Absent | Absent | Absent | Absent |
| Other findings | Normal nerve conduction studies | – | – | – | – |
| CT/MRI (very common) | Not available | MRI: hypoplastic cerebellar vermis, mega cisterna magna and bilateral symmetrical frontal lobe atrophic changes, subcortical periventricular white matter hyperintensities | Not available | CT : normal MRI: mild cerebral atrophy | Not available |
- * Abnormalities’ frequency in the previously reported cases with pathogenic PRUNE1 variants. Very common: reported in ≥80% of patients with pathogenic PRUNE1 variants; common: ≥ 50 < 80%; rare: ≥ 10 < 50; ultrarare: < 10.
3.2 Genetic Diagnosis
Our analysis identified a homozygous variant (NC_000001.10:g.150990382T > C) in the exomes of the two sequenced probands from the first family and the sequenced proband from the second family. It is absent in the exomes and genomes of 141,456 unrelated individuals in the gnomAD v2.1.1 database (Karczewski et al., 2020). This variant results in a canonical splice site alteration in PRUNE1 (NM_021222.3:c.132+2T > C). The variant was predicted to abolish the 5′ donor site according to multiple tools (NNSPLICE: 0 vs. 0.96; MaxEntScan: 0 vs. 8.23). Validation and segregation analysis showed that all five patients were homozygous, while the parents were heterozygous carriers of the variant. Other relatives were either heterozygous carriers or noncarriers. According to the American College of Medical Genetics and Genomics criteria for interpreting sequence variants (Richards et al., 2015), we classified the variant as pathogenic; we submitted it to the ClinVar database (accession: VCV000932732.1). Given the presence of a shared ROH between the two families (chr1: 150199123–151374152), we evaluated the haplotypes around PRUNE1. Microsatellite genotyping showed a shared haplotype between both families spanning roughly 1.2 mega-bases on chromosome 1, clearly confirming a founder effect (Fig. 1).
4 DISCUSSION
Here, we identified a founder variant in PRUNE1 in two families from Sudan. Five affected individuals from these families inherited a homozygous splice-donor variant that is predicted to result in a skipped exon 2. Haplotype analysis in these two families indicated the presence of a founder effect. The novelty of the PRUNE1 splice-donor variant we report in these families supports the pattern observed in the previous studies of inherited neurological disorders in this population, where we mostly observed novel variants and unique presentations (Elsayed et al., 2016a,b, 2018, 2020; Yahia et al., 2021). The affected patients had a developmental disorder, profound intellectual disability, pyramidal syndrome with prominent flexion contractures, and prominent extrapyramidal features (severe dystonia and bradykinesia). The neurodevelopmental disorder reported here in association with NM_021222.3:c.132+2T > C fits with the known phenotypic spectrum of PRUNE1 disorder, with remarkable consistency in the phenotype between the two families. However, prominent dystonia and bradykinesia are unusual disease presentations. The normal head circumference that was obtained at the time of examination in two patients from the second family (at the ages of 5 and 11 years) supports that microcephaly, although common, is not an obligatory manifestation. Serial measures of head circumference were not obtained to exclude secondary/progressive microcephaly, but previous studies showed that it usually develops at a much earlier age (Alhaddad et al., 2018; Baple et al., 2017; Imagawa et al., 2018; Karaca et al., 2015; Karakaya et al., 2017; Zollo et al., 2017). Interestingly, both microcephaly and macrocephaly were seen in patients carrying the variant NM_021222.3:c.316G > A (p.(D106N)) in a homozygous state (Imagawa et al., 2018; Milone et al., 2021).
The PRUNE1-related neurological disorder is a consequence of homozygous (most families) or compounds heterozygous (five reports) biallelic variants. These variants are characterized by considerable recurrence. A frequently detected missense variant, NM_021222.3:c.316G > A (p.(D106N)), which affects a conserved Aspartate in the DHH motif, was found in a large number of patients (about one-third of the reported families). These patients presented with a wide spectrum of typical and atypical phenotypes, including developmental and epileptic encephalopathies, PEHO syndrome (progressive encephalopathy with peripheral edema, hypsarrhythmia, and optic atrophy), as well as a spinal muscular atrophy phenotype with signs of neurogenic muscular atrophy (Chitre et al., 2018; Iacomino et al., 2018; Okur et al., 2019; Papuc et al., 2019). It is likely that NM_021222.3:c.316G > A (p.(D106N)) lies in a mutational hotspot, given the different ethnic backgrounds of the reported patients. In some populations, it has an apparent founder effect, for instance, in the Turkish population (Alhaddad et al., 2018). Another recurrent variant, NM_021222.3:c.521-2A > G, resulted in a splice site alteration (Costain et al., 2017; Hartley et al., 2019).
The novel splice-donor variant NM_021222.3:c.132+2T > C detected in the two Sudanese families reported herein is the second pathogenic PRUNE1 splice junction alteration to be identified. To date, only the recurrent splice-acceptor variant NM_021222.3:c.521-2A > G was reported, with a founder effect in the Cree population descendants (Costain et al., 2017; Hartley et al., 2019). These two variants are located in the splice junctions of two peculiar exons in PRUNE1, exons 2 and 5, respectively. Exons 2 (93 bases) and 5 (159 bases) code for amino acids in an in-frame fashion. Consequently, skipping these exons results potentially in in-frame deletions (E14_K44del and K174_G227del, respectively). In line with this hypothesis, Hartely et al. demonstrated the absence of full-length mRNA and the presence of shorter mRNA products from patients with NM_021222.3:c.521-2A > G consistent with isolated exon 5 or combined exon 4 and 5 skippings (Hartley et al., 2019). It is also known that most canonical splice alterations result in skipped exons (Anna & Monika, 2018). Activation of a cryptic splice site is another possible pathogenic mechanism for NM_021222.3:c.132+2T > C. We employed two prediction algorithms, MaxEntScan and NNSPLICE, to scan for novel splice sites. MaxEntScan (Yeo & Burge, 2004) uses the maximum entropy principle to identify splicing motifs. On the other hand, NNSPLICE (Reese et al., 1997) employs a neural network model to recognize splice sites. For both algorithms, mutated sequence scores approaching the wild-type sequence score correlate with a higher probability of novel splicing sites. Here, a possible novel 5′ splice site is predicted at position NM_021222.3:c.132+4 with a MaxEntScan score of 5.16 (vs. 8.23 for the wild-type form) and an NNSPLICE score of 0.54 (vs. 0.96 for the wild-type form). This splice site is predicted to introduce a frame shift with a consequent stop codon at the beginning of exon 3 and then nonsense-mediated mRNA decay. These predictions ideally require functional validations. However, it was not feasible to measure PRUNE1 mRNA product size in our two families to validate the predicted pathological mechanisms.
In conclusion, we identified a novel founder splice-donor variant in PRUNE1 causing a neurodevelopmental phenotype supporting the importance of splice alterations in PRUNE1-related disorders. Multiple patients from two Sudanese families presented with prominent dystonia and bradykinesia. This supports the importance of splice alterations in PRUNE1-related disorders and the potential of detecting additional occurrences of this variant in the Sudanese population.
ACKNOWLEDGMENTS
None.
AUTHORS’ CONTRIBUTIONS
LEE, AY, MK, INM, AAH, MAE, MAS, PN, AEA, HL, and GS designed the study. INM, AAH, MAE, MAS, and MHAH evaluated the patients. MK, AY, LEE, AMB, RAS, ASIAA, MM, ME, TE, HT, JA, and MRT collected the samples and relevant data and performed the experiments. MK, AY, HL, and GS interpreted the results. AEA, PN, HL, and GS supervised the study. MK, AY, and GS wrote the manuscript. All authors critically revised and approved the final variant of the manuscript. All authors greed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
FUNDING
GS was financially supported by the European Union (grant 779257 Solve-RD from the Horizon 2020 Research and Innovation Programme). HL received funding from the German Research Foundation (DFG Research Unit FOR-2715, grant Le1030/16-1). MAS was supported by Researchers Supporting Project, King Saud University, Riyadh, Saudi Arabia (project number RSP-2020/38). MK was supported by the German Academic Exchange Service (DAAD funding program number 57214224). AY was supported by a joint scholarship from the Ministry of Higher Education, Sudan, and the French embassy, Sudan.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study (not including participants’ personal information or identifiers) are available from the corresponding author upon reasonable request.




