Full-length transcriptome sequencing reveals extreme incomplete annotation of the goat genome
Abstract
Despite recent advances in generating high-quality reference genome assemblies, the genome sequences for most livestock species, including goats, are still poorly annotated. Single-molecule long-read sequencing has greatly facilitated gene annotation by obtaining full-length transcripts. In this study, we generated full-length transcriptome data for samples from abomasum (n = 2) and testicle (n = 1), using PacBio Iso-Seq technology. We further combined these data with published data from abomasum (5ZY, SRR8618141) to evaluate and improve the gene annotation of the goat genome. We identified 14.5–16.3% of novel genes per sample from the four Iso-Seq datasets. At the transcript level, 40.6% of them were novel, including 29.7% novel transcripts from known genes and 10.9% from novel genes. We further verified the expression of novel genes in four additional RNA-seq data and found that the expression level of novel genes was significantly lower than that of known genes, indicating that the lowly expressed genes tend to be missed in the current genome annotation. This study shows the superiority of full-length transcriptome data in gene annotation, and more such data are required to improve the gene annotation for goat genome and other species.
The goat, with a long history of domestication, provides economic production for humans as an important source of meat, milk and fur (Boyazoglu et al., 2005). A high-quality goat reference genome was assembled in 2017 using single-molecule sequencing, which provides a good start for the molecular breeding research and applications at the genome level of this species (Bickhart et al., 2017). The latest Ensembl release 108.1 annotated 21 361 genes with 41 794 transcripts in the goat genome compared with 19 813 coding genes of 252 477 transcripts in the human genome (Ensembl release 108.38). Given that the goat and human genomes are roughly 3 GB in size, the much lower number of annotated genes in goat suggests extreme incompleteness in the gene annotation. To investigate the genetic basis and molecular regulatory mechanisms of economically important goat traits, complete and accurate gene annotation is required in addition to high-quality genome assembly sequences.
The transcriptome is direct evidence for gene annotation, but traditional RNA-seq technology based on short-read lengths (within 150 bp) cannot directly obtain full-length transcripts and alternative splicing information. The emergence of full-length transcriptome sequencing, represented by both the PacBio and Nanopore platforms, overcomes the shortcomings of RNA-seq and allows direct access to the full-length information of transcripts (Nudelman et al., 2018). The full-length transcriptome provides an important way to improve the quality of genome annotation, which has been studied in a variety of plant and animal species (Ali et al., 2021; Feng et al., 2019; Zhang et al., 2022). Recent studies have shown that a full-length transcriptome enables the identification of novel genes and lncRNAs, the identification of full-length splice isoforms and the detection of novel alternative splicing (AS) and alternative polyadenylation events (Grabherr et al., 2011; He et al., 2022).
In order to explore the extent to which a full-length transcriptome can improve the annotation of the goat genome, we analyzed PacBio Iso-Seq data from three abomasum and one testis sample, of which three were generated in this study. By comparing with the reference genome annotation, a large number of novel genes and novel AS events were identified, necessitating the application of a full-length transcriptome for goat genome annotation.
We performed PacBio Iso-Seq for two abomasum samples (7ZY, 8ZY) and one testicle (GW) from Chinese Cashmere goats. One additional sample (5ZY) was also downloaded from the NCBI SRA database (SRR8618141) (Zheng et al., 2020). The total RNA from each sample was extracted using Trizol reagent and then sequenced using PacBio Sequel. Pacbio SMRTbell libraries were made and sequenced on two separate SMRT cells (Annoroad Gene Technology Co. Ltd). A total of 51 010 175 subreads were generated, with an average length of 1835 nt.
High-quality circular consensus sequences (CCSs) were obtained using the Isoseq3 application in the pacbio smrt analysis v6.0.0 softward (https://github.com/PacificBiosciences/IsoSeq).
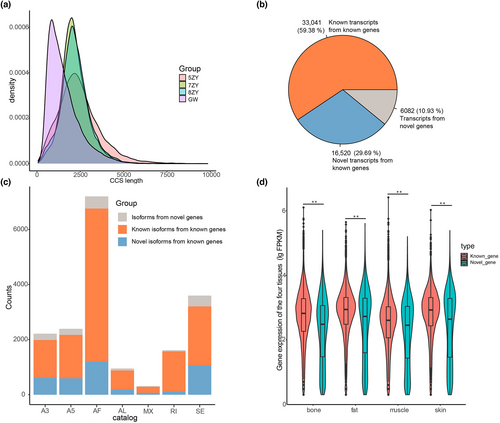
The CCSs of three abomasum samples showed a peak near 2200 bp, while the testicle sample showed a peak near 1600 bp. The CCSs of three abomasum samples displayed a peak around 2200 bp while the testicle sample showed a peak at 1600 bp. The testicle sample generated much shorter CCSs than the abomasum samples, probably owing to technical bias; however, this sample still had a similar number of CCSs above 2000 bp as the abomasum samples (Table 1, Figure 1a). Therefore, the number and average length of assembled transcripts from the testicle sample were also similar to those of the three studied abomasum samples. We classified these CCSs as full-length (FL) or non-full-length (nFL) transcript sequences using the isoseq3 software based on whether they had a 5′ or a 3′ cDNA primer and a poly(A) tail. To improve the PacBio sequencing accuracy, we chose only full-length reads. We then clustered the sequences at the isoform level using isoseq3 software, and the clustered transcripts were assembled into a complete, consistent sequence using the polish module of the software.
| Sample_id | Tissue | Number of CCSs | Number of detected genes | Number of novel genes | Number of detected transcripts | Average length of per transcript (bp) | Average number of exons per transcript |
|---|---|---|---|---|---|---|---|
| 5ZY | Abomasum | 26 073 | 9565 | 1446 | 20 782 | 2488.1 | 9.1 |
| 7ZY | Abomasum | 24 482 | 10 378 | 1560 | 21 962 | 2217.6 | 8.1 |
| 8ZY | Abomasum | 24 877 | 10 526 | 1524 | 21 198 | 2247.3 | 8.7 |
| GW | Testicle | 37 716 | 10 404 | 1697 | 22 458 | 2300.6 | 8.5 |
We identified 1 171 911 complete full-length sequences, of which 113 148 were high-quality isoforms (HQ isoforms, accuracy ≥99% and ≥2 FL reads support), and the rest were considered low-quality isoforms (LQ isoforms, accuracy <99% or only one FL read support), which were discarded. These HQ isoforms were aligned to the latest goat reference genome (ARS1, GCA_001704415.1) using minimap2 (Li, 2016). The collapse_isoforms_by_sam.py tool from cdna_cupcake software (https://github.com/Magdoll/cDNA_Cupcake) was used to filter and remove redundancy from the matching results with a minimum identity of 90% and a minimum coverage of 85%.
Novel genes and novel transcripts were detected using sqanti3_qc.py from the sqanti3 software (Tardaguila et al., 2018) from the non-redundant sequences, and then false positive transcripts were filtered using sqanti3_RulesFilter.py. A gene locus is defined as a novel gene if it does not overlap with a known gene. An isoform is defined as a novel transcript if three ‘clipping site changes or new introns (exons) emerge as compared with known transcripts’. In this way, we classified the transcripts into three categories: known transcripts from known genes; novel transcripts from known genes; and novel transcripts from novel genes. Approximately 10 000 genes were identified in each sample, of which 14.5–16.3% were novel genes, with testicular tissue having a higher number of novel genes than the others. Each gene had 2.1 transcripts on average, and the transcript harbored 8.1–9.1 exons. Among the three abomasum samples, 5ZY detected a slightly lower number of genes and transcripts than 7ZY and 8ZY, but with a longer transcript length (Table 1). Approximately 40.6% of the transcripts were novel, including 29.7% novel transcripts from known genes and 10.9% from novel genes (Figure 1b). This indicated that a large proportion of the goat genome remains poorly annotated.
All transcripts were identified using suppa2 software (https://github.com/comprna/SUPPA) for AS, including skipped exon (SE), alternative 5′ splice site (A5), alternative 3′ splice site (A5), alternative 3′ splice site (A5), alternative 3′ splice site (A3), mutually exclusive exons (MX), retained intron (RI), alternative first exons (AF) and alternative last exons (AL). AF was most prevalent, accounting for 39.4% of AS events, followed by SE (19.7%) and A5 (13.1%) (Figure 1c).
To validate the Iso-Seq sequencing data and the reliability of identified novel genes, we obtained additional genomic data from four goat tissues (bone, muscle, fat and skin), from the NCBI SRA database (PRJNA485657) (Pan et al., 2021), by quantifying their expression levels. To obtain high-quality reads, quality and length filtering of the raw sequencing reads were performed using the fastp software (Chen et al., 2018). High-quality reads were then mapped to the reference genome generated from full-length transcripts using star software (Dobin et al., 2013). The expression matrix of the annotated genes was calculated in fragments per kilobase of the exon model per million mapped fragments (FPKM) using featurecounts (Mortazavi et al., 2008). The majority (84.8%) of novel genes were found to be expressed in at least one sample with FPKM >1, suggesting that most of the identified novel genes were indeed present and demonstrated a considerable expression level. Meanwhile, for each of the four tissues, the overall expression of novel genes is significantly lower than that of known genes (Figure 1d), which could partially explain why they are currently not annotated.
This study identified abundant novel genes and novel alternative splicing sites that had not yet been annotated in the goat genome using four PacBio Iso-Seq data. The results showed that the current annotation of the goat genome is still very incomplete and far from representing the richness of the transcriptome. However, transcriptome profiling using long-read sequencing studies is still limited in livestock species (Beiki et al., 2019; Rosen et al., 2020; Warr et al., 2020; Yuan et al., 2021). Even in this study, we only have four Iso-Seq data available, which is insufficient for comprehensive gene annotation. In the future, more full-length transcriptome sequencing in various goat tissues will be needed to obtain accurate and complete genome annotation information, which will be a fundamental resource for functional genomics studies.
ACKNOWLEDGEMENTS
We thank the High-Performance Computing platform of Northwest A&F University for providing computing resources.
FUNDING INFORMATION
This study was supported by research grants from the National Natural Science Foundation of China to RL (31802027) and Major Projects of Science and Technology in Guangxi to YHC (GuikeAA18118041).
CONFLICT OF INTEREST STATEMENT
Authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data was uploaded to the NCBI SRA database with the accession numbers SRR11410765, SRR22542638 and SRR22542639.




