Identification of a missense variant in the porcine AGPAT gene family associated with intramuscular fat content through whole-genome sequencing
Abstract
The 1-acylglycerol-3-phosphate O-acyltransferases (AGPATs) are enzymes that catalyze the conversion of lysophosphatidic acid to phosphatidic acid, which is a precursor of triacylglycerol, the main fat reservoir in mammals. We used whole-genome sequencing of 205 pigs to identify 6639 genetic variants in the porcine AGPAT gene family. Of these, 166 common variants in the AGPAT5 gene had significant associations with fat content and composition traits. We preselected a missense single nucleotide polymorphism in exon 6 of AGPAT5 (rs196952262, A>G) for validation of its associations in 1034 pigs from the same Duroc line. The A allele showed a positive additive effect for intramuscular fat content (+1.12% ± 0.21, p < 0.001, for gluteus medius and +0.89% ± 0.33, p < 0.01, for longissimus). We also observed significant associations with fatty acid composition that were, at least in part, independent of the increased intramuscular fat. The A allele resulted in more monounsaturated fatty acids (+0.34% ± 0.15, p < 0.05, for longissimus) and a greater monounsaturated/polyunsaturated fatty acids ratio (+0.11 ± 0.04, p < 0.01, for gluteus medius and +0.13 ± 0.05, p < 0.05, for longissimus). The effect of the AGPAT5 variant on intramuscular fat was more noticeable in fatter pigs, and AGPAT5 interacts with other genes that affect overall fatness such as LEPR. AGPAT5 was the most expressed gene of the AGPAT family in pig skeletal muscle. This variant can be used as a marker in assisted selection for modulating pig fat deposition and fatty acid content.
INTRODUCTION
Intramuscular fat content (IMF) is related to organoleptic attributes and consumer acceptance of pork (Fernandez et al., 1999; Huff-Lonergan et al., 2002; Schwab et al., 2006). In turn, intramuscular fatty acid composition has implications for the nutritional value of pork. Although saturated (SFA) and monounsaturated fatty acids (MUFA) provide more favorable organoleptic and technological attributes than polyunsaturated fatty acids (PUFA) (Bekhit et al., 2013; Cameron et al., 2000; Wood et al., 2008; Wood & Enser, 2017), the intake of saturated fatty acids has been associated with increased risk of cardiovascular disease (Calder, 2015; Christophersen & Haug, 2011; Kapoor et al., 2021). Intramuscular fat content and fatty acid composition are especially relevant traits for consumer preferences for high-quality products (Estany et al., 2017) such as premium fresh pork and dry-cured products, and, as a consequence, they have been included in the selection objectives of pig lines aimed at this production. However, selection for IMF has been hindered by its unfavorable positive genetic correlation to backfat thickness (Newcom et al., 2005; Ros-Freixedes et al., 2013; Schwab et al., 2010; Suzuki et al., 2005), which is typically included in selection objectives of pig breeding programs as an indicator of carcass composition and growth efficiency. Similarly, the technical difficulties of routinely measuring fatty acid composition in live selection candidates and the strong correlation structure between fatty acid contents (Ros-Freixedes & Estany, 2014; Zhang et al., 2019) have limited selection for fatty acid composition. To overcome these difficulties, a lot of effort has been put into finding genetic variants that affect IMF and fatty acid composition (e.g., Ding et al., 2019; Pena et al., 2019; Puig-Oliveras et al., 2016; Ros-Freixedes et al., 2016; Won et al., 2018; Zhang et al., 2021) that can be used for marker-assisted selection.
In mammals, the main fat reservoir is in the form of triacylglycerols. Within skeletal muscle, triacylglycerols are mainly stored in the adipocytes but they can also be stored as droplets in the myocyte cytoplasm (Gardan et al., 2006; Machann et al., 2004). Triacylglycerols are primarily synthesized by sequential esterification of a glycerol backbone mediated first by glycerol 3-phosphate acyltransferase (GPAT), then by 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), and later by diacylglycerol acyltransferase (DGAT; Coleman & Lee, 2004; Figure 1). Each of these enzymes has multiple isoforms that are encoded by a family of genes. In pig, there are four annotated GPAT genes, five annotated AGPAT genes and two annotated DGAT genes.
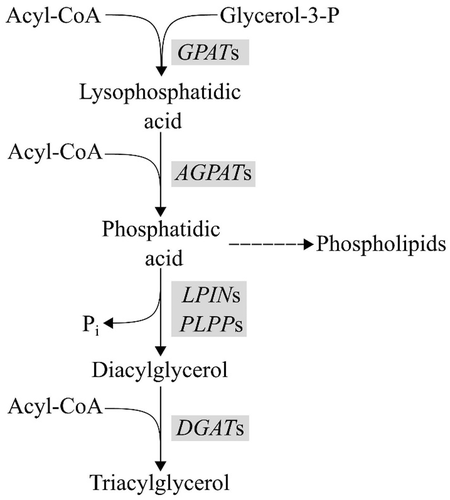
A recent study showed that a single nucleotide polymorphism (SNP) in the DGAT2 gene is associated with intramuscular palmitoleic acid content in pigs (Solé et al., 2021). Because the AGPAT genes are involved in the same metabolic pathway of triacylglycerol synthesis, those genes are plausible candidates for underlying the genetic variation of fat content and composition traits. In particular, the AGPAT enzymes catalyze the conversion of lysophosphatidic acid to phosphatidic acid. The phosphatidic group is hydrolyzed by a phosphatidic phosphatase (LPIN and PLPP) to produce diacylglycerol that later the DGAT enzyme takes as a substrate. Phosphatidic acid is not only a precursor of triacylglycerol, but is also a precursor of various glycerophospholipids and of signaling molecules involved in multiple regulatory processes, such as phosphatidylinositol, which is involved in insulin signaling (Coleman & Lee, 2004). Mutations in the AGPAT genes have been associated with body fat mass in humans (AGPAT2; Agarwal et al., 2002), mice (AGPAT1 and AGPAT2; Cortés et al., 2009; Vogel et al., 2011; Agarwal et al., 2017) and buffalo (AGPAT1 and AGPAT6; Xiaoya Ma et al., 2022), and to milk fat content in goat (AGPAT6; He et al., 2011). However, only scarce information can be found on AGPAT genes in pigs. One study in Berkshire pigs described a missense SNP (rs19695226) in the AGPAT5 gene that contributes to meat color, cooking loss and carcass temperature (Park et al., 2017).
Whole-genome sequencing is a powerful tool for variant detection. Whole-genome sequence data include rare and population-specific variants, including causative mutations (Daetwyler et al., 2014; Nicod et al., 2016; Schaid et al., 2018). The objective of this study was to use whole-genome sequencing to identify variants in the AGPAT genes and then to validate the association of preselected variants with carcass and meat quality traits, in particular backfat thickness and IMF content and composition, in pigs.
MATERIALS AND METHODS
Animals and phenotypes
A total of 1034 Duroc pigs from 182 sires and 585 dams of the same line were used in this experiment. Animals were raised in 15 batches between 2002 and 2019 following a common protocol for data recording and tissue sampling (Ros-Freixedes et al., 2013, 2016). Pigs from each batch were raised from 75 days until slaughter age (210 days, SD 9.4) under identical conditions. During this time, animals had ad libitum access to commercial feed (Esporc). At 180 days of age (SD 8.8), pigs were weighed and the backfat and loin thickness were measured at 5 cm off the midline at the position of the last rib using a portable ultrasonic scanner (Piglog 105; Frontmatec). All pigs were slaughtered in the same abattoir, where carcass weight, carcass backfat and carcass loin thickness at 6 cm off the midline between the third and fourth last ribs were recorded using an on-line ultrasound automatic scanner (AutoFOM; Frontmatec).
After chilling for approximately 24 h at 2°C, samples of the muscles gluteus medius (n = 1034) and longissimus (n = 492) were collected, vacuum-packed and stored at −20°C until required. Samples of subcutaneous fat (n = 354) were also collected and stored in the same way. The IMF content in gluteus medius and longissimus, as well as the fatty acid composition of both muscles and of subcutaneous fat, were determined in duplicate by quantitative gas chromatography (Bosch et al., 2009). Fatty acids were expressed as percentages relative to total fatty acid content. The proportion of SFA (C14:0, C16:0, C18:0 and C20:0), MUFA (C16:1n-7, C18:1n-7, C18:1n-9 and C20:1n-9), and PUFA (C18:2n-6, C18:3n-3, C20:2n-6 and C20:4n-6) were calculated.
Whole-genome sequencing
Genomic DNA from a subset of 205 pigs was isolated from gluteus medius samples using a standard protocol. The DNA samples were submitted to Centre Nacional d’Anàlisi Genòmica (CNAG-CRG) for sequencing. The short-insert paired-end libraries for the whole-genome sequencing were prepared using a PCR-free protocol using KAPA HyperPrep kit (Roche) with some modifications. In short, 0.4–1.0 μg of genomic DNA was sheared on a Covaris™ LE220-Plus focused-ultrasonicator (Covaris) in order to reach the fragment size of ~400 bp. The fragmented DNA was size-selected for the fragment size of 220–550 bp with AMPure XP beads (Agencourt, Beckman Coulter). The size-selected genomic DNA fragments were end-repaired, adenylated and ligated to adaptors with unique dual indexes and unique molecular identifiers that were compatible with the Illumina platform (Integrated DNA Technologies). The libraries were quality controlled on an Agilent 2100 Bioanalyzer with the DNA 7500 assay (Agilent) for size and quantified using a Kapa Library Quantification Kit for Illumina platforms (Roche).
The libraries were sequenced on NovaSeq6000 (Illumina) in paired-end mode with a read length of 2 × 151 + 17 + 8 bp following the manufacturer's protocol for dual indexing. Image analysis, base calling and quality scoring of the run were processed using the manufacturer's software real time analysis (RTA 3.4.4, Illumina) and followed by generation of FASTQ sequence files. A minimum of 20 Gb of sequencing data was generated per sample.
Sequencing data processing and identification of variants
DNA sequence reads were pre-processed using trimmomatic (Bolger et al., 2014) to remove adapter sequences from the reads. We mapped the reads to the reference genome Sscrofa11.1 (GenBank accession no. GCA_000003025.6; Warr et al., 2020) using the bwa-mem algorithm (Li, 2013). Duplicates were marked with picard (http://broadinstitute.github.io/picard). SNPs and short insertions and deletions (indels) were identified with the variant caller gatk haplotypecaller (GATK 3.8.0; DePristo et al., 2011; Poplin et al., 2018) using default settings. The average realized sequencing coverage was 7.9× (SD 2.4×). Variant discovery with gatk haplotypecaller was performed separately for each individual and then a joint variant set for all the individuals in each population was obtained by extracting the variant positions from all of the individuals. We retained all biallelic variants for further analyses with vcftools (Danecek et al., 2011).
Variants in the porcine AGPAT genes were retrieved including all variants in the transcription units and 500 bp upstream of the proximal promoter of the gene (for AGPAT1, SSC7, 24 204 902 to 24 213 693 bp; for AGPAT2, AEMK02000682.1, 936 421 to 1 106 715 bp; for AGPAT3, SSC13, 206 750 603 to 206 906 353 bp; for AGPAT4, SSC1, 6 772 006 to 6 883 555 bp; and for AGPAT5, SSC15, 37 800 616 to 37 853 472 bp). Note that the AGPAT2 gene is located in an unplaced scaffold in the current genome assembly. Although GPAT4 was previously named AGPAT6, we did not include it in our study because it is no longer considered a member of the AGPAT family and its ortholog showed no acylglycerol acyltransferase activity in humans (Chen et al., 2008). To determine gene expression of the members of the AGPAT family, we analyzed RNA-Seq data from muscle semimembranosus of a subset of 40 pigs from one of the batches, obtained and processed as detailed by Solé et al. (2022).
Preselection of candidate variants for fat traits
Genotyping of the candidate variant
The SNP rs196952262 (A>G) in the AGPAT5 gene (SSC15 at 37 843 344 bp) was genotyped in 1034 pigs using 5′-GTCCCTTCGAAAGCCACTGT-3′ as the forward primer and 5′-CACCAAGAATAAAGGCAACCCA-3′ as the reverse primer. Amplifications were performed by real-time PCR (QuantStudio3; Applied Biosystems, Thermo Scientific) with High-Resolution Melt analysis (Luminaris Colour HRM Master Mix; Thermo Scientific) using 20 ng of genomic DNA and 0.4 μm of each primer in a 5 μl final volume reaction. Thermocycling conditions were 95°C for 10 min, and 40 cycles of 95°C for 15 s, 60°C for 1 min, followed by a high-resolution melting curve starting with a denaturation at 95°C for 15 s, annealing at 60°C for 1 min and a slow ramp at 0.015 °C/s up to 95°C. high resolution melt software v3.1 (Applied Biosystems, Thermo Scientific) was used for the analysis of melting data and sample genotyping. All pigs were also genotyped for SNPs in genes SCD (rs80912566, C>T, on SSC14) and LEPR (rs709596309, C>T, on SSC6) following the protocols described in Estany et al. (2014), Solé et al. (2022), and Ros-Freixedes et al. (2016) respectively. In a subset of 807 pigs, the genotype for DGAT2 (rs3472408443, G>A, on SSC9) was also available (Solé et al., 2021).
Validation of the AGPAT5 variant
The effect of the AGPAT5 rs196952262 SNP genotype on production traits (body weight, carcass weight, backfat thickness, loin thickness and IMF) and fatty acid composition was estimated using a model with the same fixed effects as above (with 15 batches) plus the genotype for AGPAT5 (GG, AG and AA). The effect of the genotype was tested using the F-statistic. Multiple pairwise comparisons among AGPAT5 genotypes were tested with the Tukey HSD test. The additive and dominant effects of the AGPAT5 SNP were also estimated by replacing the genotype with two covariates coded as (−1, 0, 1) and (0, 1, 0) for the GG, AG and AA genotypes respectively. The IMF content was added as a covariate to test whether the effect of the AGPAT5 SNP on fatty acid composition was due to changes in fat content. Finally, we also tested the interaction of the AGPAT5 SNP with LEPR and SCD, as well as with DGAT2, by adding this latter SNP to previous models. All the analyses were performed using the statistical package jmp pro 15 (SAS Institute Inc.).
RESULTS
Preselection of candidate variants for fat traits
Our analysis of the genetic variation of the AGPAT gene family revealed that AGPAT5 contained the strongest candidate variants for fat content and fatty acid composition traits. We found large differences in the number of variants located in the transcription unit of the genes of the AGPAT family (Table 1). In total, 6639 variants were detected across the 205 pigs, 2220 of which had minor allele frequencies equal to or greater than 0.20 (referred to here as ‘common’ variants). Of these, 171 were prioritized as candidate variants because they showed associations with p-values ≤0.001 for at least one of the tested traits. Most of these candidate variants (166 out of 210 common variants) were located on AGPAT5, only one (out of 120 common variants) was located on AGPAT2, and four (out of 442 common variants) on AGPAT4. The five candidate variants in AGPAT2 and AGPAT4 were intronic.
| Gene | Total called variants | Common variants | Candidate variants |
|---|---|---|---|
| AGPAT1 | 55 | 0 | 0 |
| AGPAT2 | 1469 | 120 | 1 |
| AGPAT3 | 2045 | 1448 | 0 |
| AGPAT4 | 2722 | 442 | 4 |
| AGPAT5 | 348 | 210 | 166 |
| Total | 6639 | 2220 | 171 |
- Note: Common variants are variants with a minor allele frequency ≥0.2. Candidate variants are variants that showed an association with at least one trait (p ≤ 0.001).
A detailed summary of all of the results for all 348 variants in AGPAT5 is provided in Table S1 and Figure S1. Although we applied a restrictive minor allele frequency threshold, 114 of the 138 variants that were removed by this filter had actually a minor allele frequency below 0.05, of which 108 were intronic. This filter also removed a few variants that were in the promoter (one), in exon 8 (one; missense) and in the 3′-UTR (four), but their minor allele frequency was below 0.05 and no significant associations were detected for these variants, possibly in part because of the small sample size of the less frequent genotype. The remaining 210 common variants located in AGAPT5 were grouped in four different haplotype blocks (Table S1). Haplotype block 1, which included 151 variants, contained most variants with statistically significant associations with the studied traits, while haplotype blocks 2–4 were much shorter (2–58 variants each) and largely depleted of variants with statistically significant associations. Of the 210 common variants detected in AGPAT5, only 11 were in the promoter, 5′-UTR and 3′-UTR and exonic regions of the gene (Table 2), all of which were in moderate linkage disequilibrium (r2 > 0.7) with SNP223 (rs196952262). Three of these variants, including SNP223, were located in exons and were predicted to be missense mutations.
| Variant | Position (bp) | Location | Major/minor allele | MAFa | Association study with IMFb | Association study with all traitsc | Predicted consequenced | Linkage disequilibrium with SNP223 (r2) |
|---|---|---|---|---|---|---|---|---|
| SNP2 | 37 800 993 | Promoter | C/T | 0.266 | 1.06 | 2.75 | – | 0.85 |
| SNP4 | 37 801 250 | 5′-UTR | A/C | 0.263 | 1.06 | 4.23 | – | 0.79 |
| SNP223 | 37 843 344 | Exon 6 | G/A | 0.259 | 1.80 | 3.38 | Missense (2/2) | – |
| SNP333 | 37 850 594 | Exon 8 | G/C | 0.263 | 1.20 | 2.88 | Missense (1/2) | 0.77 |
| SNP334 | 37 850 596 | Exon 8 | G/T | 0.263 | 1.20 | 2.88 | Missense (1/2) | 0.77 |
| INDEL338 | 37 851 301 | 3′-UTR | G/GAAAC | 0.278 | 1.33 | 3.99 | – | 0.76 |
| SNP339 | 37 852 012 | 3′-UTR | C/A | 0.281 | 0.57 | 3.61 | – | 0.70 |
| SNP342 | 37 852 233 | 3′-UTR | C/T | 0.231 | 0.64 | 3.26 | – | 0.70 |
| SNP345 | 37 852 886 | 3′-UTR | C/T | 0.278 | 0.71 | 4.30 | – | 0.76 |
| INDEL347 | 37 853 314 | 3′-UTR | G/GT | 0.276 | 1.21 | 3.74 | – | 0.80 |
| INDEL348 | 37 853 443 | 3′-UTR | G/GA | 0.272 | 1.38 | 3.13 | – | 0.78 |
- a MAF, minor allele frequency.
- b p-value, expressed as –log(p-value), of the association of the variant with intramuscular fat content (IMF).
- c p-value, expressed as –log(p-value), of the most significant association of the variant with any of the studied traits. For all displayed variants, the most significantly associated trait was C16:0, which has a phenotypic correlation of 0.47 with IMF.
- d Predicted consequence on the annotated gene transcripts. In parentheses, the number of transcripts affected over the total number of transcripts.
Only the missense variant SNP223 was predicted to be missense for both annotated transcripts of the gene, while the other two were missense for only one of the two transcripts. SNP223 was also the missense variant that showed the highest association with the tested traits (p ≤ 0.001). SNP223 produces a replacement of an isoleucine by a valine. Owing to the similarity of those amino acids, the mutation is predicted to be tolerated based on its Sorting Intolerant From Tolerant (SIFT) score (Ng & Henikoff, 2003). The sequence of AGPAT5 is highly conserved between species; however, the Genomic Evolutionary Rate Profiling (GERP) conservation score (Cooper et al., 2005) indicated that SNP223 was a highly variable position. Taking into account these results, together with the fact that SNP223 (rs196952262) was previously reported in Berkshire pigs as associated with meat quality traits (Park et al., 2017), we preselected this variant as a tag SNP for further validation.
Validation of the AGPAT5 variant
The effect of the AGPAT5 SNP rs196952262 on fat content was validated using data from 1034 pigs (Table 3). The AGPAT5 SNP was associated with production traits at 180 days of age, with the A allele showing a positive additive effect on backfat thickness (+0.60 mm ± 0.27, p < 0.05) and a negative effect on loin thickness (−0.85 mm ± 0.35, p < 0.05). These effects were less clearly detected when measured on carcass around 4 weeks later and only loin thickness showed a similar trend (−0.96 mm ± 0.52, p < 0.10). The A allele also was positively associated with IMF, both in gluteus medius (+1.12% ± 0.31, p < 0.001) and in longissimus (+0.89% ± 0.33, p < 0.01).
| Trait | AGPAT5 genotype | Additive effect1 | Dominant effect | ||||
|---|---|---|---|---|---|---|---|
| AA (n = 65) | AG (n = 358) | GG (n = 611) | a | p-value | d | p-value | |
| Live measurements (180 days) | |||||||
| Body weight, kg | 110.5 ± 1.5 | 110.4 ± 0.6 | 109.5 ± 0.5 | 0.51 ± 0.78 | 0.51 | 0.38 ± 0.95 | 0.69 |
| Backfat thickness, mm | 20.5 ± 0.5 | 19.7 ± 0.2 | 19.3 ± 0.2 | 0.60 ± 0.27 | 0.02 | −0.20 ± 0.32 | 0.53 |
| Loin thickness, mm | 43.2 ± 0.7a | 45.0 ± 0.3b | 44.9 ± 0.2b | −0.85 ± 0.35 | 0.02 | 0.98 ± 0.43 | 0.02 |
| Carcass measurements (210 days) | |||||||
| Carcass weight, kg | 96.9 ± 1.2 | 98.0 ± 0.52 | 96.8 ± 0.4 | 0.01 ± 0.63 | 0.98 | 1.15 ± 0.78 | 0.14 |
| Backfat thickness, mm | 24.0 ± 0.4 | 24.3 ± 0.18 | 23.8 ± 0.1 | 0.09 ± 0.22 | 0.66 | 0.36 ± 0.27 | 0.19 |
| Loin thickness, mm | 42.2 ± 1.0ab | 42.8 ± 0.43a | 44.1 ± 0.3b | −0.96 ± 0.52 | 0.07 | −0.32 ± 0.64 | 0.62 |
| Lean content, % | 40.4 ± 0.7 | 40.1 ± 0.3 | 40.9 ± 0.2 | −0.29 ± 0.36 | 0.43 | 0.57 ± 0.45 | 0.21 |
| Intramuscular fat, % dry matter | |||||||
| Muscle gluteus medius | 20.42 ± 0.59a | 18.85 ± 0.26b | 18.18 ± 0.20b | 1.12 ± 0.31 | <0.001 | −0.45 ± 0.39 | 0.24 |
| Muscle longissimus2 | 15.65 ± 0.64a | 14.45 ± 0.31ab | 13.86 ± 0.26b | 0.89 ± 0.33 | 0.007 | −0.31 ± 0.41 | 0.46 |
- Note: Bold font indicates statistical significance (p ≤ 0.05).
- a,bWithin traits, means with different superscripts indicate statistically significant differences (p ≤ 0.05).
- 1 Additive allele substitution of G by A.
- 2 Sample size was 38, 187 and 267 for AA, AG, and GG respectively.
Differences in intramuscular fatty acid composition were also detected across AGPAT5 genotypes. The A allele had a consistent effect in both muscles (Tables 4 and 5). The A allele had a positive additive effect on MUFA (+0.35% ± 0.13, p < 0.01, in gluteus medius and +0.50% ± 0.16, p < 0.01, in longissimus), mainly owing to its effect on C18:1n-9. Also, the A allele showed a negative effect on PUFA (−0.45% ± 0.13, p < 0.001, in gluteus medius and −0.54% ± 0.18, p < 0.01, in longissimus), with decreased values for C18:2n-6, C18:3n-3 and C20:4n-6. As a result, the A allele showed a positive effect on the MUFA/PUFA ratio (0.22 ± 0.04, p < 0.001, in gluteus medius and 0.34 ± 0.09, p < 0.001, in longissimus). Since the effects of the A allele on MUFA and PUFA counteract each other, the AGPAT5 genotype did not influence the SFA/(MUFA+PUFA) ratio. We observed no differences in fatty acid composition for subcutaneous fat (Table S2).
| Trait | AGPAT5 genotype | Additive effect1 | Dominant effect | ||||
|---|---|---|---|---|---|---|---|
| AA (n = 65) | AG (n = 358) | GG (n = 611) | a | p-value | d | p-value | |
| Fatty acid, % | |||||||
| SFA | 37.92 ± 0.24 | 37.83 ± 0.11 | 37.73 ± 0.08 | 0.10 ± 0.13 | 0.46 | 0.01 ± 0.16 | 0.95 |
| C16:0 | 24.39 ± 0.15 | 24.43 ± 0.07 | 24.31 ± 0.05 | 0.05 ± 0.08 | 0.60 | 0.08 ± 0.10 | 0.42 |
| C18:0 | 11.73 ± 0.11 | 11.56 ± 0.05 | 11.59 ± 0.04 | 0.07 ± 0.06 | 0.22 | −0.10 ± 0.07 | 0.16 |
| MUFA | 50.37 ± 0.25a | 49.75 ± 0.11b | 49.66 ± 0.09b | 0.35 ± 0.13 | 0.006 | −0.26 ± 0.16 | 0.11 |
| C16:1n-7 | 3.70 ± 0.07 | 3.72 ± 0.03 | 3.66 ± 0.02 | 0.02 ± 0.04 | 0.61 | 0.03 ± 0.05 | 0.47 |
| C18:1n-7 | 4.26 ± 0.04 | 4.30 ± 0.02 | 4.29 ± 0.02 | −0.01 ± 0.02 | 0.47 | 0.03 ± 0.03 | 0.28 |
| C18:1n-9 | 42.02 ± 0.19a | 41.66 ± 0.10ab | 41.50 ± 0.09b | 0.26 ± 0.11 | 0.01 | −0.09 ± 0.13 | 0.48 |
| PUFA | 11.71 ± 0.24a | 12.41 ± 0.11b | 12.61 ± 0.08b | −0.45 ± 0.13 | <0.001 | 0.24 ± 0.16 | 0.13 |
| C18:2n-6 | 9.43 ± 0.19a | 9.89 ± 0.08ab | 10.03 ± 0.06b | −0.30 ± 0.10 | 0.002 | 0.16 ± 0.12 | 0.18 |
| C18:3n-3 | 0.56 ± 0.01 | 0.57 ± 0.01 | 0.58 ± 0.01 | −0.01 ± 0.01 | 0.10 | 0.01 ± 0.01 | 0.30 |
| C20:4n-6 | 1.20 ± 0.07a | 1.37 ± 0.03b | 1.46 ± 0.02c | −0.13 ± 0.03 | <0.001 | 0.045 ± 0.04 | 0.30 |
| Fatty acid ratio | |||||||
| MUFA/SFA | 1.35 ± 0.02 | 1.33 ± 0.01 | 1.34 ± 0.01 | 0.01 ± 0.01 | 0.55 | −0.01 ± 0.01 | 0.44 |
| MUFA/PUFA | 4.57 ± 0.09a | 4.23 ± 0.04b | 4.14 ± 0.03b | 0.22 ± 0.04 | <0.001 | −0.13 ± 0.06 | 0.03 |
| SFA/(MUFA+PUFA) (×10) | 6.16 ± 0.06 | 6.14 ± 0.03 | 6.11 ± 0.02 | 0.02 ± 0.03 | 0.49 | 0.00 ± 0.04 | 0.95 |
- Note: Bold font indicates statistical significance (p ≤ 0.05).
- Abbreviations: C16:0, palmitic acid; C18:0, stearic acid; C16:1n-7, palmitoleic acid; C18:1n-7, vaccenic acid; C18:1n-9, oleic acid; C18:2n-6, linoleic acid; C18:3n-3, linolenic acid; C20:4n-6, arachidonic acid; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids.
- a–cWithin trait, means with different superscripts indicate statistically significant differences (p ≤ 0.05).
- 1 Additive allele substitution of G by A.
| Trait | AGPAT5 genotype | Additive effect1 | Dominant effect | ||||
|---|---|---|---|---|---|---|---|
| AA (n = 38) | AG (n = 187) | GG (n = 267) | a | p-value | d | p-value | |
| Fatty acid, % | |||||||
| SFA | 39.56 ± 0.34 | 39.51 ± 0.16 | 39.48 ± 0.14 | 0.04 ± 0.17 | 0.82 | −0.01 ± 0.22 | 0.97 |
| C16:0 | 25.40 ± 0.20 | 25.29 ± 0.10 | 25.29 ± 0.18 | 0.06 ± 0.10 | 0.59 | −0.06 ± 0.13 | 0.65 |
| C18:0 | 12.46 ± 0.17 | 12.45 ± 0.08 | 12.41 ± 0.07 | 0.02 ± 0.09 | 0.80 | 0.02 ± 0.11 | 0.87 |
| MUFA | 51.22 ± 0.32a | 50.50 ± 0.16ab | 50.22 ± 0.13b | 0.50 ± 0.16 | 0.002 | −0.22 ± 0.20 | 0.29 |
| C16:1n-7 | 4.00 ± 0.09 | 3.92 ± 0.04 | 3.97 ± 0.03 | 0.02 ± 0.04 | 0.72 | −0.06 ± 0.06 | 0.31 |
| C18:1n-7 | 4.34 ± 0.06 | 4.33 ± 0.03 | 4.34 ± 0.03 | −0.00 ± 0.03 | 0.97 | −0.01 ± 0.04 | 0.77 |
| C18:1n-9 | 42.07 ± 0.29a | 41.73 ± 0.15ab | 41.30 ± 0.14b | 0.39 ± 0.16 | 0.01 | 0.05 ± 0.21 | 0.82 |
| PUFA | 9.21 ± 0.34a | 9.99 ± 0.17ab | 10.30 ± 0.14b | −0.54 ± 0.18 | 0.002 | 0.23 ± 0.22 | 0.31 |
| C18:2n-6 | 7.18 ± 0.24a | 7.73 ± 0.12ab | 7.93 ± 0.10b | −0.37 ± 0.13 | 0.003 | 0.18 ± 0.16 | 0.26 |
| C18:3n-3 | 0.34 ± 0.01 | 0.37 ± 0.01 | 0.37 ± 0.01 | −0.01 ± 0.01 | 0.03 | 0.01 ± 0.01 | 0.14 |
| C20:4n-6 | 1.31 ± 0.09a | 1.46 ± 0.05ab | 1.57 ± 0.04b | −0.13 ± 0.05 | 0.006 | 0.02 ± 0.06 | 0.7 |
| Fatty acid ratio | |||||||
| MUFA/SFA | 1.31 ± 0.02 | 1.29 ± 0.01 | 1.29 ± 0.01 | 0.01 ± 0.01 | 0.22 | −0.01 ± 0.01 | 0.57 |
| MUFA/PUFA | 5.78 ± 0.18a | 5.26 ± 0.09b | 5.11 ± 0.07b | 0.34 ± 0.09 | <0.001 | −0.18 ± 0.12 | 0.12 |
| SFA/(MUFA+PUFA) (×10) | 6.59 ± 0.09 | 6.57 ± 0.04 | 6.57 ± 0.04 | 0.01 ± 0.05 | 0.82 | −0.01 ± 0.06 | 0.91 |
- Note: Bold font indicates statistical significance (p ≤ 0.05).
- Abbreviations: C16:0, palmitic acid; C18:0, stearic acid; C16:1n-7, palmitoleic acid; C18:1n-7, vaccenic acid; C18:1n-9, oleic acid; C18:2n-6, linoleic acid; C18:3n-3, linolenic acid; C20:4n-6, arachidonic acid; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids.
- a,bWithin trait, means with different superscripts indicate statistically significant differences (p ≤ 0.05).
- 1 Additive allele substitution of G by A.
To discard the possibility that the discovery set was underlying these significant associations, the associations of the AGPAT5 SNP with fat content and composition traits were confirmed in the validation data after excluding the 205 pigs used for discovery (Tables S3–S5), although the statistical significance decreased, probably owing to the reduced sample size. The rest of the analyses were performed with the whole dataset to maximize the sample size as part of a sequential validation process (where we accumulated all available data) rather than a cross-validation.
With the exception of MUFA and C18:1n-9, the effects of the A allele on fatty acid composition were mostly due to changes in fat content, since they were not detected when adjusted for IMF (Tables S6, S7). The effects of the A allele on MUFA, which was driven mainly by its effect on C18:1n-9, was in part independent of IMF, although this was more evident in longissimus (+0.34% ± 0.15, p < 0.05, for MUFA and +0.27% ± 0.15, p < 0.10, for C18:1n-9) than in gluteus medius (+0.21% ± 0.12, p < 0.10, for MUFA and +0.16% ± 0.10, p = 0.11, for C18:1n-9). As a result, the A allele displayed a positive additive effect on the MUFA/PUFA ratio in both gluteus medius (+0.11 ± 0.04, p < 0.01) and longissimus (+0.13 ± 0.05, p < 0.05), even after adjusting for IMF.
We detected a significant interaction between AGPAT5 and LEPR for IMF (Table S8). Although the effect of AGPAT5 on IMF showed the same trend within each LEPR genotype, the additive effect of the A allele of AGPAT5 was larger in the LEPR-TT pigs (+1.47% ± 0.72, p < 0.05), which are fatter, than in the LEPR-CC or CT pigs (+0.99% ± 0.34, p < 0.01). We also confirmed the effect of AGPAT5 on IMF in the DGAT2-GG pigs (Table S9), although statistical evidence for its effect within the other DGAT2 genotypes was limited to assess the interaction between these two genes. We detected some significant interactions for fatty acid composition between AGPAT5 and genes LEPR (Table S8) and SCD (Table S10), but they did not show clear patterns that were biologically meaningful. There were no interactions between AGPAT5 and DGAT2 for fatty acid composition (Table S9).
DISCUSSION
In this study, we identified variants in the family of AGPAT genes using whole-genome sequencing and preselected a candidate variant for fat content and composition traits. Using a large number of individuals, we validated the effect of the rs196952262 SNP as a tag SNP for the haplotype in AGPAT5. In the following we will discuss: (1) the suitability of whole-genome sequencing for preselecting candidate variants in the AGPAT gene family associated to traits of interest; and (2) the effect of the AGPAT5 SNP on fat content and composition.
Preselection of candidate variants in the AGPAT gene family
Whole-genome sequencing is a powerful tool for detecting large numbers of variants that could be associated with complex traits. However, the identification of casual variants remains challenging because the high density of variants that are in high linkage disequilibrium hinders the disentangling of the causal variant. We found the strongest evidence of association with fat content and composition traits for AGPAT5. In contrast, we did not find any candidate variant in AGPAT1 or AGPAT3, and the few candidate variants in AGPAT2 and AGPAT4 were discarded because they were located in introns. A typical criterion for prioritizing candidate variants is to limit the search to coding and promoter regions, since the prediction of the potential effects of variants located in non-coding regions from DNA sequence is not straightforward (Johnsson & Jungnickel, 2021). However, in some instances non-coding variants, which may have regulatory functions, have been proposed as candidate variants (Ryan et al., 2012; Solé et al., 2021; Van Laere et al., 2003), while variants in coding regions have often been found to have a small impact on complex traits (Koufariotis et al., 2018; Xiang et al., 2019). Although mutations in AGPAT2 that affected triacylglycerol synthesis and storage have been described in humans (Agarwal et al., 2002), we did not detect any orthologous polymorphism in pigs with a significant association with the studied traits.
The results for AGPAT5 are consistent with the fact that this gene is the most expressed of the AGPAT gene family in skeletal muscle in pigs. Using data from a previous RNA-Seq experiment (Solé et al., 2022), we found that AGPAT5 and AGPAT2 are the most expressed genes in muscle semimembranosus in pig (Figure S2). Those results are also in line with previous gene expression data reported in pig skeletal muscle (Freeman et al., 2012). In contrast, expression analysis in humans showed that AGPAT1 and AGPAT2 were highly expressed in adipocytes and skeletal muscle (Agarwal et al., 2002; Prasad et al., 2011), while in mice AGPAT1 and AGPAT2 and AGPAT4 showed the highest overall expression (Vergnes et al., 2006). Although our expression data were insufficient to detect any potential expression differences between AGPAT5 genotypes (Figure S3), the fact that in pigs AGPAT5 is the most expressed gene of the AGPAT family, in contrast to human and mice, supports the contribution of this isoform in the conversion of lysophosphatidic into phosphatidic acid in pig skeletal muscle, and hence, in triacylglycerol synthesis. If these results were confirmed, they could help understand differences in the metabolic pathways of fat deposition of pigs compared with other mammals.
We preselected the rs196952262 SNP as a tag variant for the AGPAT5 gene based on several criteria. Coincidentally, the same SNP was previously reported to be associated with meat quality in Berkshire pigs (Park et al., 2017) for traits such as meat color, cooking loss and carcass temperature, albeit not for backfat thickness. In particular, these authors found that the A allele was associated with a lighter pork color (greater CIE L* parameter). Although these authors did not analyze the IMF, the lighter color associated with the A allele is compatible with greater levels of IMF (Schwab et al., 2006; Suárez-Mesa et al., 2021), which adds another layer of evidence in support of this variant from an independent population.
Although the SIFT and GERP scores of rs196952262 indicated that this variant was not predicted as deleterious or highly conserved across species, this was not unexpected and it should not be taken as compelling evidence against the relevance of rs196952262 in artificial settings because variants that affect fat content and composition traits may not be under purifying selection as much as other variants that are more directly related to fitness in the wild. Similarly, we took rs196952262 as the tag variant for haplotype block 1 and, although other variants in this haplotype block (e.g. intronic variants) could also be causal, our method does not allow us to distinguish between them owing to linkage disequilibrium. Additional studies in independent populations where these variants segregate and functional assays are required to elucidate causality.
A limitation of our methodology is that we discarded a lot of variants with minor allele frequencies below 0.2, although most discarded variants had minor allele frequencies below 0.05 (Table S1). Although variants that are more likely to affect traits are typically expected to have low minor allele frequencies owing to selection pressure, it would be unlikely that we could estimate their effects with enough accuracy with 205 sequenced pigs and then validate them. Moreover, these rare variants typically contribute low percentages of genetic variance and, therefore, little to the selection response. In contrast, our method targeted other variants with intermediate minor allele frequencies that may not have been affected by such a selection pressure and may segregate in the population.
Effect of the AGPAT5 SNP on fat content and composition
We validated the association of rs196952262 SNP from AGPAT5 on fat content and composition. This association had not been identified in previous analyses in Duroc. We observed that the A allele had a positive additive effect on backfat thickness at 180 days and on IMF in both muscles. Alterations of the gene activity led to lower triacylglycerol synthesis, and therefore to a reduced fat accumulation (Cortés et al., 2009). Although we expected to observe the same additive effect on carcass backfat thickness, we were not able to detect this effect, probably because of differences in age and measuring methods for live and carcass backfat thickness, but collectively our results indicate an effect on overall carcass fatness. The rs196952262 SNP explained 1.2 and 1.4% of the genetic variance for backfat and loin thickness at 180 days respectively, but much less at 210 days (0.03 and 0.6% respectively). The rs196952262 SNP had a higher impact on IMF than on backfat thickness. The SNP explained 2.3 and 2.5% of the genetic variance for IMF in gluteus medius and longissimus respectively. Moreover, we observed differences in fatty acid composition that were specific to intramuscular fat and did not affect subcutaneous fat, which might further indicate a lower impact on backfat thickness. The A allele also had a negative effect on loin thickness and thus, attention should be paid to any unfavorable correlated responses in carcass lean content.
The A allele segregated at high frequency in Iberian pigs (0.81, n = 18), which is consistent with the body fatness that characterizes this breed, and at intermediate frequency in much leaner breeds such as Pietrain (0.47, n = 28) and a Large White × Landrace crossbred (0.50, n = 49). Despite this, it segregated at lower frequencies in other heavily marbled breeds such as Duroc, both the population studied here (0.24) and in two other Duroc populations (0.15, n = 26, and 0.25, n = 20) and Berkshire (0.21; Park et al., 2017). The lower frequency in Duroc pigs indicated that selection assisted by this marker could increase the population average IMF in gluteus medius by up to +1.7% if the A allele was fixed.
The rs196952262 SNP was also associated with intramuscular fatty acid composition. The effects that we observed for AGPAT5 on fatty acid composition were consistent with the faster accumulation of SFA and MUFA relative to PUFA as fat reserves grow (De Smet et al., 2004; Ros-Freixedes et al., 2016; Zhang et al., 2019). However, we found some evidence of an effect of AGPAT5 on fatty acid composition that was, at least in part, independent of its effect on fat content. In particular, AGPAT5 showed an IMF-independent effect on MUFA, mainly driven by C18:1n-9. The rs196952262 SNP explained 0.5% of the genetic variance for MUFA in gluteus medius and 1.0% in longissimus. In humans, functional studies of the AGPAT proteins showed that both AGPAT3 and AGPAT5 had a higher affinity for C18:1n-9 as a substrate, which could explain our results (Prasad et al., 2011). As a consequence of this, we found significant differences for the MUFA/PUFA ratio between genotypes and the SNP could be used as a marker for selecting pork with a higher monounsaturated fatty acid profile.
The effect of AGPAT5 on IMF may become more noticeable in fatter pigs. Thus, AGPAT5 showed significant interactions with genes that affect overall fatness, such as LEPR. In turn, this can produce changes in fatty acid composition, to the extent that composition indirectly reflects changes in fat content. However, there were no clear interactions between AGPAT5 and SCD despite the fact that both genes affect MUFA. Even though the AGPAT5 gene takes part in the same metabolic pathway as DGAT2, in a previous study we found no effect of DGAT2 on IMF (Solé et al., 2021) and therefore an interaction effect of these two genes on IMF seems unlikely. In contrast, DGAT2 has a specific effect on intramuscular C16:1n-7 (Solé et al., 2021), but we did not detect any effect of AGPAT5 on this fatty acid.
In conclusion, our analysis of the genetic variation in the sequence of the genes of the AGPAT family revealed that AGPAT5 contained the strongest candidate variant for fat content and fatty acid composition traits. The A allele of the rs196952262 variant is associated with increased IMF and MUFA but it is negatively associated with loin thickness. The association between AGPAT5 and IMF is more noticeable in fatter pigs and, therefore, AGPAT5 could interact with genes that affect overall fatness, such as LEPR. Although further studies would be needed before the causality of the variant can be confirmed, this variant can be used as a selection marker for modulating pig fat deposition and the fatty acid composition of pork.
ACKNOWLEDGEMENTS
We acknowledge the personnel at Selección Batallé for their cooperation for the recording of on-farm data and sample collection. We gratefully acknowledge Pilar Sopeña from the Animal Breeding group, University of Lleida, for laboratory assistance.
FUNDING INFORMATION
This research was supported by the Spanish Ministry of Science, Innovation & Universities and the EU Regional Development Funds (grant RTI2018-101346-B-I00). EM is recipient of a UdL-Santander Predoc scholarship.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ETHICAL APPROVAL
The pigs used in this study were raised and slaughtered in commercial units complying with the regulations and good practice guidelines on the protection of animals kept for farming purposes, during transport and slaughter (Royal Decree 37/2014, Spain). Tissue samples used in this study were collected at the slaughterhouse. The Ethical Committee on Animal Experimentation of the University of Lleida approved all experimental procedures.
Open Research
DATA AVAILABILITY STATEMENT
The dataset for the replication of the main results of this study is available from CORA Research Data Repository with doi 10.34810/data167. Sequence variants are available from the European Variation Archive of EMBL with the accession number ERZ6113288. RNA-Seq data are available from NCBI-GEO with the accession number GSE183909.




