Taking advantage from phenotype variability in a local animal genetic resource: identification of genomic regions associated with the hairless phenotype in Casertana pigs
Summary
Casertana is an endangered autochthonous pig breed (raised in south-central Italy) that is considered to be the descendant of the influential Neapolitan pig population that was used to improve British breeds in the 19th century. Casertana pigs are characterized by a typical, almost complete, hairless phenotype, even though a few Casertana pigs are normal haired. In this work, using Illumina PorcineSNP60 BeadChip data, we carried out a genome-wide association study and an FST analysis with this breed by comparing animals showing the classical hairless phenotype (n = 81) versus pigs classified as haired (n = 15). Combining the results obtained with the two approaches, we identified two significant regions: one on porcine chromosome (SSC) 7 and one on SSC15. The SSC7 region contains the forkhead box N3 (FOXN3) gene, the most plausible candidate gene of this region, considering that mutations in another gene of the same family (forkhead box N1; Foxn1 or FOXN1) are responsible for the nude locus in rodents and alopecia in humans. Another potential candidate gene, rho guanine nucleotide exchange factor 10 (ARHGEF10), is located in the SSC15 region. FOXN3 and ARHGEF10 have been detected as differentially expressed in androgenetic and senescent alopecia respectively. This study on an autochthonous pig breed contributes to shed some light on novel genes potentially involved in hair development and growth and demonstrates that local animal breeds can be valuable genetic resources for disclosing genetic factors affecting unique traits, taking advantage of phenotype variability segregating in small populations.
Local animal genetic resources might be characterized by specific and inheritable phenotypes with relevant importance for current or potential future use in breeding programs or for many other purposes, including the definition of new biological models or to understand mechanisms of biological adaptations to different environments (Leroy et al. 2016).
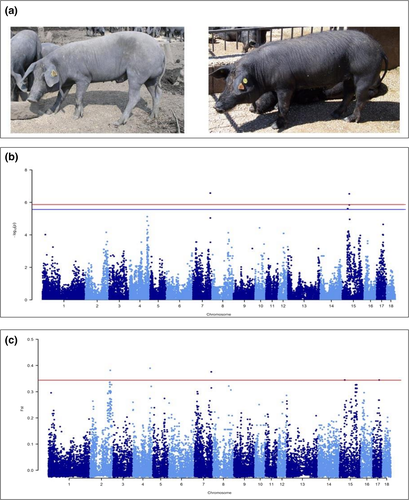
Casertana is an endangered autochthonous pig breed raised mainly in south-central Italy, accounting for about 100 boars and sows currently registered to its herd book (ANAS 2016). Casertana pigs are usually raised in extensive or semi-extensive systems to produce niche pork products. This local breed is considered the descendant of the influential Neapolitan breed of the late 18th and 19th centuries that was used to improve British pig populations from which several modern commercial breeds were derived (Porter 1993). Casertana pigs are characterized by a black or grey coat colour, wrinkled skin, forward ears, two goat-like wattles (not always present) and a typical, almost complete, hairless phenotype not related to the age of the animals. This latter characteristic is also reported in one of its local names, i.e. Pelatella (which means plucked or bald). Despite the hairless phenotype being the characteristic phenotype of this breed, the Casertana population shows some variability for this trait, including animals having an almost complete absence of hairs (hairless; the most common pigs) to a few animals having abundant hair (normal-haired pigs; Fig. 1a). The hairless phenotype is also present in other pig breeds such as the Creole hairless Mexican breed (also known as Pelón Mexicano) and the black hairless Iberian strains, including the Guadyerbas strain maintained as an isolated population (Toro et al. 2000; Lemus-Flores et al. 2001). Casertana and all these other hairless pigs seem historically connected through the exchange of pig genetic material related to commercial activities in the 18th and 19th centuries (Porter 1993), suggesting a potential common origin of the hairless phenotype.
Hairless or hairlessness in pigs can be better described as hypotrichosis or congenital deficiency of hairs, as animals classified as ‘hairless’ usually show a small number rather than a complete absence of hairs. Roberts & Carroll (1931) were the first authors to report a possible inheritance model for this hypotrichotic condition in Mexican pigs, suggesting the presence of a monogenic autosomal factor with a recessive mutated h allele that could give the hairless phenotype when homozygous. Pigs homozygous for the wild type allele H might be normal haired, whereas heterozygous Hh pigs might show an intermediate phenotype. This early study was not followed up with any other genetic investigations on the hairless condition in pigs. More recently, variability in the porcine hairless gene (known as HR, lysine demethylase and nuclear receptor corepressor), located on porcine chromosome (SSC) 14, was evaluated in a candidate gene approach to study the hairless phenotype in Iberian pigs, but no association with this trait was reported (Fernández et al. 2003, 2006). Mutations in the HR gene have been shown to impair hair growth in different mammals (i.e. Stoye et al. 1988; Ahmad et al. 1998; Finocchiaro et al. 2003). A high number of other genes in humans and rodents have been implicated in abnormal hair development and hypotrichosis (Shimomura & Christiano 2010; Ramot & Zlotogorski 2015), making it impractical to use a candidate gene approach to successfully identify polymorphisms associated with the hairless phenotype in pig populations.
In this work, with the aim to restrict the number of potential causative genes involved in the hypotrichotic phenotype in pigs, we carried out a genome-wide association study (GWAS) and a genome-wide FST analysis in the Casertana breed by comparing animals showing the classical hairless phenotype (n = 81; 35 males and 46 females) versus pigs classified as haired (n = 15; seven males and eight females; a quite rare phenotype in this breed), without any distinction between possible different hair levels, which could not be recorded precisely in outdoor animals. The Casertana breed offers a unique opportunity to investigate this phenotype that is segregating within the same population. This is one of the first population-based genome-wide studies with a local pig breed and is useful not only to characterize a breed-specific trait but also to obtain basic biological information that could be important in better defining an interesting animal model for alopecia or related phenotypes in humans (Shimomura 2012).
Blood or hair roots were collected from all these Casertana pigs raised in six different farms located in the Campania and Molise regions (south central Italy), each farm having from five to 49 pigs with unknown relationships. A two-tailed chi-square analysis with Yates correction confirmed that the occurrence of the observed phenotypes was not associated with the sex of the sampled animals (P > 0.10). Extracted DNA was used for genotyping with the Illumina PorcineSNP60 BeadChip v.2, interrogating 61 565 single nucleotide polymorphisms (SNPs). Genotyping data were processed with plink 1.9 software (Chang et al. 2015) using the following criteria to filter SNPs: call rate greater than 0.9, minor allele frequency greater than 0.01 and Hardy-Weinberg equilibrium P > 0.001. A total of 36 533 autosomal SNPs, assigned to a unique position in the Sscrofa11.1 version genome, were used for multidimensional scaling (MDS) obtained with plink 1.9 software (Chang et al. 2015) to evidence distance relationships among the animals of the investigated cohort. The obtained MDS plot showed some structures in the analysed pigs not well defined and did not clearly separate the two Casertana groups (i.e. hairless and haired; Fig. S1).
The GWAS was then carried out using the filtered SNPs by applying the univariate mixed model of GEMMA to be able to correct for population relatedness and possible clusterisation (Zhou & Stephens 2012). The centered relatedness matrix calculated from SNP genotypes was included in the model to correct for population stratification. The genomic inflation factor (λ) and quantile–quantile (Q–Q) plot, obtained with genabel (Aulchenko et al. 2007), is shown in Fig. S2. The Manhattan plot produced in this GWAS is shown in Fig. 1b. Relevant data reported in this work have been submitted to the Zenodo digital repository. At the P < 0.05 Bonferroni corrected level (Pnominal value < 1.37E-06), three SNPs were significant, whereas at the P < 0.1 Bonferroni corrected threshold (Pnominal value = 2.74E-06) three additional SNPs were suggestively significant (Table 1). Two of these SNPs were located on SSC7 (170.17 kb apart) and four on SSC15, in two distinct regions of approximately 1.14 Mb and 338.61 kb.
| SSC | SNP | Position | GWAS Pnominal value | FST value | Annotated genes |
|---|---|---|---|---|---|
| 2 | ALGA0016212 | 134 598 604 | – | 0.381 | – |
| 4 | INRA0016870 | 113 277 535 | – | 0.390 | – |
| 7 | INRA0028322 | 111 520 662 | 2.68E-07 | 0.376 | LOC106504536, PSMC1, EFCAB11, NRDE2, CALM1, TDP1, KCNK13, FOXN3 |
| 7 | ALGA0044817 | 111 690 832 | 2.68E-07 | 0.376 | |
| 15 | MARC0009352 | 33 679 138 | 2.45E-06 | 0.345 | LOC110257074, CLN8, KBTBD11, DLGAP2, LOC106509653, ARHGEF10, LOC106506202, CSMD1, MYOM2 |
| 15 | ALGA0084906 | 34 793 592 | 2.45E-06 | 0.345 | |
| 15 | H3GA0044265 | 44 006 149 | 3.00E-07 | – | – |
| 15 | INRA0049225 | 44 344 760 | 1.43E-06 | – | – |
| 17 | DRGA0016747 | 41 675 886 | – | 0.345 | – |
| 17 | H3GA0049027 | 41 643 251 | – | 0.345 | – |
- SSC, porcine chromosome number.
- For the overlapping regions among the two approaches, annotated genes near the SNPs (±500 kb from the first to the last SNP of the region) were reported (Sscrofa11.1 genome version). The candidate genes that could be involved in the hair phenotype are indicated in bold. P, FST and annotated genes are reported only for the SNPs and regions for which both P and FST values exceeded the indicated thresholds.
The FST analysis was performed on the same dataset using plink 1.9 software (Chang et al. 2015). Missing SNPs were imputed using beagle 3.3.2 software (Browning & Browning 2009). The Manhattan plot of the FST analysis is shown in Fig. 1c. The top 0.9998 SNPs of the percentile distribution (FST = 0.345) were considered as the most divergent across the comparison and therefore were retained for subsequent evaluation (Table 1). Eight SNPs were above the selected threshold: one on SSC4, one on SSC2, two on SSC7 (170.17 kb apart), two on SSC15 (1.14 Mb apart) and two on SSC17 (32.00 kb apart).
The comparison among GEMMA and FST genome-wide analyses identified two overlapping regions encompassing two SNPs on SSC7 and two SNPs on SSC15 that constituted the 1.14-Mb region previously mentioned (Table 1). Eight and nine genes were annotated in the SSC7 and SSC15 regions respectively (in windows ±500 kb from the first and the last SNPs; Table 1). The most plausible candidate gene in the SSC7 region was the forkhead box N3 (FOXN3) gene (position: 111 036 492–111 454 106 bp), which is 66.56 kb away from INRA0028322 (one of the two most significant SNPs in the GWAS; Table 1). This gene has a role in the regulation of hepatic glucose utilization (Karanth et al. 2016), craniofacial development (Samaan et al. 2010) and growth and migration of colon cancer cells (Dai et al. 2017). The FOXN3 gene was also found to be differentially expressed in a case–control study for androgenetic alopecia in humans (Mirmirani & Karnik 2010). Forkhead box proteins constitute a family of transcription factors involved in embryo and fetal development and function of adult organisms (Hannenhalli & Kaestner 2009). The list of this group of proteins has about 50 members in mammals, divided into 19 subfamilies indicated with letters from A to S (Jackson et al. 2010; Benayoun et al. 2011). Within the N subfamily, forkhead box N1 (FOXN1) regulates keratin gene expression and the gene (Foxn1) is responsible for the nude locus in rodents (Flanagan 1966; Meier et al. 1999). Mutations in this gene determine hairlessness, alopecia and other pleiotropic effects in mice and rats (Nehls et al. 1994) as well as congenital alopecia, nail dystrophy and primary T-cell immunodeficiency in humans (Frank et al. 1999). Therefore, considering the phylogenetic relationships and the partially conserved domains between the FOXN1 and FOXN3 genes (Benayoun et al. 2011), it seems plausible that FOXN3 might have conserved regulatory functions similar to those of FOXN1 that could explain the effect of this SSC7 chromosome region on the hairless phenotype in Casertana pigs. This indication might contribute to the understanding of the involvement of forkhead box proteins in hair development and, if confirmed by functional studies, adds another candidate gene to the list of those potentially involved in alopecia and baldness.
No strong candidate gene could be identified in the SSC15 region. A possible candidate could be the rho guanine nucleotide exchange factor 10 (ARHGEF10) gene. ARHGEF10 is involved in neural morphogenesis and connectivity and in the regulation of small RhoGTPases (Verhoeven et al. 2003). The ARHGEF10 gene has been reported to be differentially expressed in a case–control study of senescent alopecia in human (Mirmirani & Karnik 2010), supporting, to some extent, its possible role in the hairless phenotype in the Casertana breed. According to the available functional information, no other gene in the two identified regions might be involved in hair or follicle development or phenotypes similar to the hairless condition we investigated.
The combination of the GWAS and FST results with the annotated gene functions was useful for drafting a possible biological explanation for the hairless phenotype in Casertana pigs and for identifying significant regions, excluding other regions that reached or were close to the defined thresholds in one or the other genome-wide investigation methods derived by several confounding factors that could not be better managed in our study (i.e. genetic drift, population structure, ascertain bias of the SNP chip tool). However, the results obtained in this breed, even if based on a small group of pigs with the normal-haired phenotype (which is a quite rare in this breed) in contrast with the hairless group, seems to support the presence of more than one locus affecting this trait. A few of the associated genomic regions contain candidate genes that, based on their function or inferred function, may be involved in the hypotricotic condition of the Casertana pigs, with the hypothesis that this trait might be more complex than previously suggested.
This study provides another contribution to the genetic characterization of morphological traits in pigs that have been reported to describe breed-specific phenotypes (i.e. ear size and coat colours) in other autochthonous populations (i.e. Ren et al. 2011; Fontanesi et al. 2016). This work demonstrates that endangered animal genetic resources can be investigated to disclose genetic factors affecting unique traits, taking advantage of phenotype variability segregating within a small population. Other investigations are needed to refine these results obtained in Casertana and to evaluate if the hairless condition in other pig breeds is derived by the same genetic factors identified in this study.
Competing interests
The authors declare that they do not have competing interests. Data reported in this work can be shared after signature of an agreement on their use with University of Bologna.
Acknowledgements
This work has received funding from the Italian Ministry of Agriculture, Food and Forestry (MiPAAF) under the project INNOVAGEN and from the European Union's Horizon 2020 research and innovation programme under grant agreement No 634476 (Project acronym: TREASURE). The content of this paper reflects only the authors' view, and the European Union Agency is not responsible for any use that may be made of the information it contains.




