Using genetic information from candidate gene and genome-wide association studies in risk prediction for alcohol dependence
Abstract
Family-based and genome-wide association studies (GWAS) of alcohol dependence (AD) have reported numerous associated variants. The clinical validity of these variants for predicting AD compared with family history information has not been reported. Using the Collaborative Study on the Genetics of Alcoholism (COGA) and the Study of Addiction: Genes and Environment (SAGE) GWAS samples, we examined the aggregate impact of multiple single nucleotide polymorphisms (SNPs) on risk prediction. We created genetic sum scores by adding risk alleles associated in discovery samples, and then tested the scores for their ability to discriminate between cases and controls in validation samples. Genetic sum scores were assessed separately for SNPs associated with AD in candidate gene studies and SNPs from GWAS analyses that met varying P-value thresholds. Candidate gene sum scores did not exhibit significant predictive accuracy. Family history was a better classifier of case-control status, with a significant area under the receiver operating characteristic curve (AUC) of 0.686 in COGA and 0.614 in SAGE. SNPs that met less stringent P-value thresholds of 0.01–0.50 in GWAS analyses yielded significant AUC estimates, ranging from mean estimates of 0.549 for SNPs with P < 0.01 to 0.565 for SNPs with P < 0.50. This study suggests that SNPs currently have limited clinical utility, but there is potential for enhanced predictive ability with better understanding of the large number of variants that might contribute to risk.
Introduction
Alcohol dependence (AD) is a complex psychiatric condition that is influenced by both genetic and environmental factors. It has a lifetime prevalence of 12.5% and affects 4–5% of individuals at any given time in the United States (Hasin et al. 2007). It also impacts other diseases (Hasin et al. 2007). Based on twin studies, AD has an estimated heritability of around 50–60% (Kendler et al. 1992; Heath et al. 1997). Survey studies suggest that there may be interest in genetic counseling and testing to determine risk for AD (Gamm, Nussbaum & Biesecker 2004). More than half of individuals surveyed who had at least one first-degree relative with AD reported that they would undergo a genetic test to determine their own risk for AD if one were available. Many of them believed that testing would lead to better prevention or treatment and help assess their own children's risk (Gamm et al. 2004). Current risk assessment for AD does not include genetic testing for common variants; the predictive value of genetic testing has yet to be determined. This research reveals a need for the careful evaluation of the clinical utility of genetic information for predicting AD.
There has been a recent emergence of direct-to-consumer (DTC) personal genomics testing for many multi-factorial disorders, including addiction, despite limited information about the clinical validity and utility of genetic variants associated with these disorders (Mathews, Hall & Carter 2012). Public interest in genetic testing may be due in part to a misunderstanding of how predictive genetics can be for complex disorders (Lawrence & Appelbaum 2011). Genetic counseling for AD is designed to help individuals understand, manage and cope with risk so that they have less anxiety and a greater sense of mastery over this disorder, although the actual level of control may be modest (Peay et al. 2008). Current assessment of risk for AD involves taking a detailed personal and family history of clinical and subclinical features for AD, possible co-occurring conditions in the family, and environmental risk factors (Peay et al. 2008). Empiric risk estimates derived from population-based family studies are also included as risk assessment tools for AD. However, risk estimates from a population sample may not be applicable for a specific individual due to differences in genetic and environmental backgrounds. Furthermore, empiric risk may not be available for families with multiple psychiatric phenotypes or across all family relationships (Austin & Peay 2006). Genetic information specific to the individual may therefore provide more accurate recurrence risk assessments than empiric risk estimates.
Previous efforts to study risk prediction for complex disorders have assessed the predictive ability of genetic sum scores based on number of risk alleles that have been associated with a particular disorder. The ability of a test to distinguish between individuals with and without a disease is typically assessed based on the test's sensitivity, or the proportion of individuals with the condition who have a positive result on the test, and specificity, or the proportion of individuals without the condition who test negative. A frequent measure of clinical validity is the receiver operating characteristic (ROC) curve, which plots the sensitivity versus 1-specificity for every cut-off of a continuous predictor to distinguish between the presence and absence of a disease diagnosis. The area under the ROC curve (AUC) for a continuous predictor corresponds to the probability that an individual with the disease would have a higher predictor score than an individual without the disease, and therefore reflects the proportion of individuals classified correctly as cases or controls. An AUC of 0.5 means that the predictor can accurately classify 50% of individuals, or no greater than chance, whereas an AUC of 1.0 means that the predictor can correctly classify 100% of individuals. An AUC of 0.80 is generally accepted as a target cut-off for screening and 0.99 for diagnosis (Janssens et al. 2006). Simulation studies that we have conducted suggest that if all genetic contributions are included in a prediction model for AD, given AD's heritability of around 50%, there is the potential for AUCs approaching 0.80 to be reached with genetic information alone (Maher et al., in preparation).
ROC curve analyses of prior complex diseases have shown modest predictive ability of genetic sum scores, with AUCs of 0.54 for diabetes for a genetic risk score created based on previously associated variants (Talmud et al. 2010) to 0.65 corresponding to the 3% of variance in schizophrenia risk explained by a risk score created based on a large number of single nucleotide polymorphisms (SNPs) that met less-stringent P-value thresholds in genome-wide association studies (GWAS) (Purcell et al. 2009; Jostins & Barrett 2011). Most of the genetic variants contributing to AD have small effect sizes. This, along with the fact that AD has both genetic and environmental risk factors, means that any one SNP alone is not expected to be a good predictor of AD. This study aims to explore the aggregate impact of multiple genetic variants with small effect sizes on risk prediction in order to test whether known genetic contributions to AD can be an effective predictor.
The Collaborative Study on the Genetics of Alcoholism (COGA) is a National Institutes of Health-sponsored project aimed at identifying genes that contribute to alcohol-related outcomes. In COGA, we have conducted a series of analyses aimed at understanding the underlying genetic architecture of AD (Zlojutro et al. 2011). Here, we couple this knowledge with a clinical evaluation of the information captured by currently available genetic information in risk prediction for AD. COGA has previously reported positive family-based association results for AD using a high-density family sample. Many of these genes have also been associated with AD in other studies (Table 1). We created additive genetic sum scores based on risk alleles of associated SNPs in these genes. We then compared the sum score with family history in its ability to discriminate between cases and controls for AD in a subset of the COGA sample that is independent of the gene-finding family sample and in a subset of independent individuals in the Study of Addiction: Genes and Environment (SAGE) GWAS sample. Finally, we explored the clinical validity of results from genome-wide association analyses.
| Study | Gene | Replication |
|---|---|---|
| Edenberg et al., 2004 | GABRA2 | Covault et al., 2004; Fehr et al., 2006; Lappalainen et al., 2005; Soyka et al., 2008; Enoch et al., 2006; Drgon et al., 2006 |
| Dick et al., 2004 | GABRB3 and GABRG3 | Noble et al., 1998; Song et al., 2003 GABRB3 |
| Wang et al., 2004 | CHRM2 | Luo et al., 2005a |
| Hinrichs et al., 2006 | TAS2R16 | |
| Wang et al., 2009 | CHRNA5 | Saccone et al., 2007 |
| Xuei et al., 2006 | PDYN and OPRK1 | Williams et al., 2007; Gerra et al., 2007 |
| Edenberg et al., 2006 | ADH genes: ADH4, ADH1A, ADH1B | Luo et al., 2005b; Guindalini et al., 2005 |
| Edenberg et al., 2008 | NFKB1 | |
| Foroud et al., 2008 | TACR3 | |
| Dick et al., 2008 | ACN9 | |
| Dick et al., 2007b | ANKK1/DRD2 |
- COGA, Collaborative Study on the Genetics of Alcoholism.
Materials and Methods
Sample and measures
COGA family-based association analysis sample
COGA is a large-scale multi-center family study with 10 collaborative sites across the United States. The sample consists of families containing probands meeting both DSM-III-R and Feighner criteria for AD ascertained since 1989 from outpatient and in-patient alcohol treatment centers at six sites across the United States: Indiana University, State University of New York Health Science Center, University of Connecticut, University of Iowa, University of California/San Diego, and Washington University in St Louis. Families were interviewed using a poly-diagnostic instrument, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA), which assesses Feighner, DSM-III-R, DSM-IV and ICD-10 criteria for major psychiatric disorders (American Psychiatric Association 1987, 2000; Feighner et al. 1972). More than 1300 probands with AD have been recruited. Unaffected subjects were defined as individuals who drank but did not meet criteria for AD or illicit substance dependence. A subset of the COGA sample was identified as a group of high-density families with three or more first-degree relatives who met lifetime criteria for AD. The institutional review boards from all of the participating institutions approved the study (Edenberg et al. 2008; Wang et al. 2009).
SNPs included in this analysis were selected from nine COGA papers reporting family-based association analyses for AD using individuals from the high-density subset (Table 1). The number of individuals included varied across studies: association analyses that encompassed all ancestries ranged from 2139 to 2310 individuals from 262 families; 35 of these families, comprising a total of 298 individuals, are of African-American (AA) ancestry. Analyses conducted in the European American (EA) subset ranged from 1172 to 1923 individuals from 217–219 families. Genotyping for these individuals is described in detail in the original COGA papers. Briefly, SNPs within and flanking candidate genes were selected from public databases including dbSNP (http://www.ncbi.nlm.nih.gov/SNP), HapMap (http://www.hapmap.org) and LocusLink (http://www.ncbi.nlm.nih.gov/gene). Genotyping was done using a modified single nucleotide extension reaction, with allele detection by mass spectrometry (Sequenom MassArray system, Sequenom, San Diego, CA, USA). SNPs were in Hardy Weinberg Equilibrium. Genotypes were checked for Mendelian inheritance using programs including PEDCHECK. USERM13 was used to calculate marker allele frequencies and heterozygosities (Edenberg et al. 2008).
COGA GWAS sample
A case-control sample of 1945 phenotyped subjects was selected from the larger COGA sample for GWAS. Cases had a lifetime diagnosis of AD by DSM-IV criteria. Controls reported consuming alcohol but did not have a diagnosis of AD or alcohol abuse by any of the diagnostic criteria assessed by SSAGA and did not meet diagnostic criteria for dependence on cocaine, marijuana, opioids, sedatives or stimulants. Controls could not share a known common ancestor with a case and were preferentially selected to be above the age of 25 years.
Genotyping was completed using the Illumina Human 1 M DNA Analysis BeadChip at the Center for Inherited Disease Research. Additional details on the COGA GWAS sample can be found in Edenberg et al. (2010).
SAGE GWAS sample
The SAGE is part of the Gene Environment Association Studies initiative of the National Human Genome Research Institute to identify genetic contributions to addiction through large-scale GWAS. The entire SAGE sample consists of 4121 cases and unrelated controls from subsets of three large studies on addiction: the Family Study of Cocaine Dependence (FSCD), the Collaborative Genetic Study of Nicotine Dependence (COGEND), and COGA. All cases in SAGE have a DSM-IV lifetime diagnosis of AD. Controls were exposed to alcohol. Some controls met criteria for nicotine dependence based on the Fagerström Test for nicotine dependence, but none met criteria for a DSM-IV lifetime dependence diagnosis for alcohol, marijuana, cocaine, opiates or other drug. Genotyping for the SAGE GWAS sample was completed using the Illumina Human 1 M DNA Analysis BeadChip. The institutional review boards at all participating sites granted approval for data collection in COGA, COGEND and FSCD in the SAGE sample. Additional details on the SAGE GWAS sample can be found in Bierut et al. (2010).
Family history measures
Family history information for the COGA GWAS sample was obtained for both cases and controls as a dichotomous ‘yes’/‘no’ variable for any existence of a family history of AD, as reported by the subject. The SAGE GWAS sample included a ‘yes’/‘no’ variable about history of AD in specifically the proband's mother and father. The presence or absence of family history was used as a binary variable in order to reflect clinical scenarios in which an individual is asked whether or not he or she has a family history of AD.
Data analysis
Analysis for this study was broken down into two parts, distinguished by whether SNPs were selected from family-based candidate gene association studies or from case-control GWAS analyses (Fig. 1). In the first part of this study, SNPs that were previously associated with AD in candidate gene studies in the COGA high-density family-based association sample were used to create a genetic risk score to assess prediction of AD in independent individuals from the COGA and SAGE GWAS samples. The second part of the study assessed the discriminatory, or predictive, accuracy of SNP panels selected from GWAS results using varying ‘significance’ criteria. We controlled for allele frequency and linkage disequilibrium (LD) pattern differences across ethnicities by assessing risk scores in just the EA subsets in both parts of the study. In order to select independent discovery and validation samples, individuals from the COGA GWAS EA sample independent of the COGA family-based association sample were used to assess predictive accuracy of candidate gene sum scores. The FSCD and COGEND portions of the SAGE GWAS EA sample were extracted for use as a sample independent of COGA. Table 2 summarizes characteristics of the samples used in both study parts. Discriminatory accuracy of genetic sum scores and family history was measured using ROC curve analysis in SPSS/PASW v17.0 (SPSS Inc., Chicago, IL, USA) and the caTools package (Tuszynski 2011) in R v2.12.2 (R Foundation, Vienna, Austria).
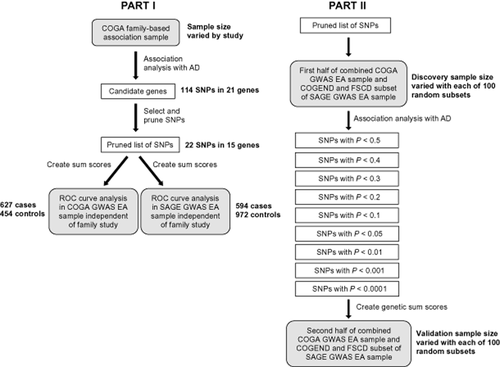
Study overview. Gray boxes show samples used for each step of analyses. White boxes display the selection criteria for single nucleotide polymorphisms (SNPs) at each step. AD = alcohol dependence; COGA = Collaborative Study on the Genetics of Alcoholism; COGEND = Collaborative Genetic Study of Nicotine Dependence; EA = European American; FSCD = Family Study of Cocaine Dependence; GWAS = genome-wide association studies; ROC = receiver operating characteristic; SAGE = Study of Addiction: Genes and Environment
| Characteristic | COGA family sample | COGA GWAS EA sample | FSCD EA subset of SAGE GWAS sample | COGEND EA subset of SAGE GWAS sample | ||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| Sample size | 909 | 1291 | 846 | 552 | 275 | 241 | 335 | 702 |
| Sex | ||||||||
| Male, n (%) | 601 (66.1) | 434 (33.6) | 590 (69.7) | 151 (27.4) | 150 (54.5) | 103 (42.7) | 171 (51.0) | 174 (24.8) |
| Female, n (%) | 308 (33.9) | 857 (66.4) | 256 (30.3) | 395 (71.6) | 125 (45.5) | 138 (57.3) | 164 (49.0) | 528 (75.2) |
| Age, years | ||||||||
| Mean, SD | 37.6 ± 12.3 | 42.5 ± 15.9 | 46.6 ± 12.2 | 41.8 ± 11.0 | 33.0 ± 8.9 | 34.0 ± 9.2 | 36.9 ± 6.1 | 37.1 ± 6.8 |
| Range | 18–80 | 17–91 | 18–79 | 18–78 | 18–52 | 18–54 | 25–61 | 25–65 |
| AD symptom count | ||||||||
| Mean, SD | 5.3 ± 1.5 | 0.8 ± 1.1 | 5.6 ± 1.4 | 0.1 ± 0.3 | 5.5 ± 1.5 | 0.7 ± 0.8 | 4.4 ± 1.3 | 0.9 ± 0.8 |
| Family historya | ||||||||
| Negative, n (%) | 0 (0) | 0 (0) | 213 (34.0) | 323 (71.1) | 140 (50.9) | 208 (86.3) | 247 (73.7) | 605 (86.2) |
| Positive, n (%) | 909 (100) | 1291 (100) | 414 (66.0) | 131 (28.9) | 135 (49.1) | 33 (13.7) | 88 (26.3) | 97 (13.8) |
- a Family history represents parental history in FSCD and COGEND and any family history in COGA GWAS sample. Family history in COGA GWAS is presented here for 1081 individuals independent of COGA family sample. AD = DSM-IV alcohol dependence; COGA = Collaborative Study on the Genetics of Alcoholism; COGEND = Collaborative Genetic Study of Nicotine Dependence; EA = European American; FSCD = Family Study of Cocaine Dependence; GWAS = genome-wide association studies; SAGE = Study of Addiction: Genes and Environment; SD = standard deviation. Case-control status is based on DSM-IV diagnosis of AD. All cases and controls are unrelated in GWAS samples and related in the COGA family sample.
Part I: family-based SNP panel
SNP selection
Several criteria were used to select SNPs for the genetic sum score. An initial list of 114 SNPs across 21 genes was generated based on prior association with AD (Table 1). SNPs associated only with early onset AD were not included in the list so that SNPs in the candidate gene panel would be applicable to the wide range of ages of individuals in the COGA and SAGE validation samples. Because assessment of clinical validity was to be performed in EA individuals, SNPs that were associated only in the AA subset were removed from the list. Forty-two of the SNPs showing association in the original papers (Table 1) were present on the Illumina Human 1 M DNA Analysis BeadChip. Because we wanted to include SNPs that were captured on the current GWAS arrays, we used proxy SNPs for SNPs that were not genotyped on the arrays rather than used imputed SNPs or removed the SNPs altogether. Proxy SNPs on the Illumina chip with an r2 > 0.70 were found for 32 additional SNPs based on LD calculations in the HapMap CEU data using Haploview (Barrett et al. 2005) and PLINK v1.07. An additional 32 SNPs did not have proxies. Seven of these SNPs had proxies in the list of COGA family sample SNPs for which proxy SNPs existed on the Illumina chip, based on LD calculations using Haploview. The final list contained 81 SNPs.
SNP pruning
In order that genes with a large number of associated SNPs in high LD were not disproportionately represented in the risk panel, we generated a list of semi-independent SNPs for the panel and removed SNPs with an r2 greater than 0.50. LD estimations used for pruning the SNPs were based on the HapMap Phase 3 CEU data using the PLINK v1.07 LD function. Selection of which SNP of a pair of correlated SNPs to remove depended on a ranked list of SNPs based on the level of significance from the family-based association results and how closely the SNP on the Illumina chip matched the original family-based SNP. Table 3 summarizes the list of SNPs after pruning. Pruning resulted in a set of 22 SNPs in 15 genes, with several genes pruned out primarily due to correlations among the ADH SNPs.
| SNP | Status | Gene | COGA family study P value | MAF Fam | MAF COGA | MAF SAGE | Risk allele |
|---|---|---|---|---|---|---|---|
| rs10499934 | In_sample | ACN9 | 0.003 | 0.23 | 0.22 | 0.23 | A |
| rs12671685 | In_sample | ACN9 | 0.027 | 0.11 | 0.12 | 0.11 | A |
| rs7794886 | In_sample | ACN9 | 0.006 | 0.35 | 0.36 | 0.35 | T |
| rs4147531 | In_sample | ADH1A | 0.007 | 0.43 | 0.46 | 0.47 | C |
| rs1229982 | In_sample | ADH1B | 0.048 | 0.22 | 0.20 | 0.19 | T |
| rs1126672 | In_sample | ADH4 | 0.010 | 0.29 | 0.28 | 0.29 | C |
| rs17115439 | In_sample | ANKK1 | 0.096 | 0.33 | 0.32 | 0.32 | C |
| rs680244 | In_sample | CHRNA5 | 0.114 | 0.42 | 0.41 | 0.42 | G |
| rs1799978 | In_sample | DRD2 | 0.168 | 0.06 | 0.05 | 0.05 | G |
| rs279858 | In_sample | GABRA2 | 0.010 | 0.38 | 0.42 | 0.42 | A |
| rs1897356 | In_sample | GABRB3 | 0.020 | 0.17 | 0.15 | 0.15 | C |
| rs16918941 | In_sample | OPRK1 | 0.023 | 0.06 | 0.06 | 0.07 | G |
| rs6985606 | In_sample | OPRK1 | 0.004 | 0.48 | 0.50 | 0.48 | T |
| rs997917 | In_sample | OPRK1 | 0.011 | 0.27 | 0.29 | 0.27 | C |
| rs1997794 | In_sample | PDYN | 0.011 | 0.37 | 0.36 | 0.35 | C |
| rs2235749 | In_sample | PDYN | 0.010 | 0.27 | 0.27 | 0.26 | A |
| rs6045819 | In_sample | PDYN | 0.038 | 0.10 | 0.12 | 0.12 | G |
| rs11722288 | In_sample | TACR3 | 0.022 | 0.29 | 0.29 | 0.29 | G |
| rs3762894 | In_sample | ADH4 | 0.050 | 0.16 | 0.15 | 0.16 | C |
| rs1391175 | Use_proxy rs13120165 | GABRG1 | 0.036 | 0.06 | 0.03 | 0.03 | A |
| rs3097490 | Use_proxy rs1571281 | GABRG3 | 0.137 | 0.44 | 0.44 | 0.46 | G |
| rs324640 | Use_proxy rs324649 | CHRM2 | 0.038 | 0.43 | 0.42 | 0.42 | T |
- ‘Status’ indicates whether or not the SNP was genotyped directly on the Illumina 1 M SNP chip or a proxy SNP was used. The SNP numbers are SNPs from candidate gene studies, with proxy SNPs indicated as such in the ‘status’ column. The COGA family-based association P values from our rerun analyses are listed. ‘MAF Fam’ shows the minor allele frequency of the SNP in the COGA family-based candidate gene association sample. ‘MAF COGA’ and ‘MAF SAGE’ correspond to the MAF in the COGA and SAGE GWAS samples, respectively. The risk allele corresponds to the GWAS alleles matched by allele frequency to the risk allele in the family-based candidate gene association sample. COGA = Collaborative Study on the Genetics of Alcoholism; SAGE = Study of Addiction: Genes and Environment; SNP = single nucleotide polymorphism.
Genetic risk scores
Sum scores were created using the—score option in PLINK v1.07 (Purcell et al. 2007). The number of risk alleles was added and then divided by the number of non-missing genotypes to create a normalized allele count for each individual. Because odds ratios associated with the risk alleles varied across family-based analyses in COGA and replication studies, an additive score was created without weighting alleles by effect size. The risk allele in the SAGE and COGA samples was determined by matching by frequency with alleles that were associated with AD in the family sample.
Association analysis of panel SNPs with AD
Sum scores were tested for association with DSM-IV AD in the case-control COGA and SAGE samples using logistic regression with sex as a covariate in COGA, and sex, age quartiles and study site as covariates in SAGE. The models were selected to follow the methods used in the previously reported primary COGA and SAGE GWAS analyses (Bierut et al. 2010; Edenberg et al. 2010). In addition to testing the sum scores, the individual SNPs contributing to the scores were also tested for association with AD in the sample used for prediction. All association analyses were completed in the case-control samples using logistic regression using an additive model in PLINK v1.07 for both the EA subset of the sample and the entire sample, including individuals of non-EA ancestry. Association analyses in the entire GWAS samples that included individuals of non-EA ancestry included molecularly derived principal components factor covariates, PC1 and PC2, distinguishing primarily between European and African ancestry.
Part II: GWAS results from varying P-value thresholds
Sample selection
The FSCD and COGEND subset of the SAGE EA sample was combined with the COGA GWAS EA sample, and then split randomly in half so that each half contained 50% of cases and 50% of controls. In order to account for chance effects, this subsetting procedure was performed 100 times to obtain 100 subsamples in which analyses were completed. The combined sample included 2951 individuals, comprising of 1456 cases and 1495 controls. Controls who endorsed three or more symptoms for DSM-IV AD, but did not cluster within a 12-month period, were removed from the combined sample, as these individuals may still represent genetic risk (N = 49).
SNP pruning
The LD-based pruning function in PLINK v1.07 was used to prune the 1 041 983 SNPs genotyped in the combined sample before association analyses were performed. The SNPs were pruned at r2 < 0.50 using a sliding window of 50 base pairs shifted by five base pairs following each pruning step.
Association analyses
Association was performed using logistic regression with sex and site covariates distinguishing between the three study sites using an additive model in PLINK v1.07. Figure 1 shows the P-value thresholds used to select SNPs from association results in the first half of the sample.
Genetic sum scores
Because both GWAS samples had the same SNPs genotyped, and were confirmed to share the direction of the genotyped strand, GWAS results were matched directly by allele. Genetic sum scores were created for autosomal SNPs composed of the total number of minor alleles for each SNP carried by each individual, so that homozygotes for the risk allele had a score of 2. Each SNP allele count was weighted by the natural log of the odds ratio for each minor allele, and then the sum of the weighted allele count was divided by the number of non-missing genotypes for each individual using PLINK v1.07. The P values associated with the AUCs for these sum scores were calculated based on the Wilcoxon rank-sum test using R v2.12.2.
Results
Family-based SNP panel
Association of candidate gene sum scores and individual SNPs from candidate genes with AD
The sum scores for the panel of SNPs were not associated with AD in the COGA or SAGE samples. Logistic regression results for individual SNPs within the panels from the COGA family-based study are shown in Table 4. Logistic regression P values of the expanded panel of SNPs prior to LD-based pruning resulted in a greater number of SNPs that met nominal association levels for AD, and are summarized in Supporting Information Table S1.
| CHR | SNP | Gene | P-val COGA EA | P-val COGA All | P-val SAGE EA | P-val SAGE All |
|---|---|---|---|---|---|---|
| 4 | rs13120165 | GABRG1 | 0.104 | 0.842 | 0.388 | 0.569 |
| 4 | rs279858 | GABRG1 | 0.681 | 0.429 | **0.024 | **0.017 |
| 4 | rs1126672 | ADH4 | *0.073 | *0.069 | 0.922 | 0.449 |
| 4 | rs3762894 | ADH4 | 0.510 | 0.128 | 0.592 | 0.501 |
| 4 | rs4147531 | ADH1A | 0.571 | 0.940 | 0.806 | 0.508 |
| 4 | rs1229982 | ADH1B | 0.104 | 0.337 | 0.604 | 0.594 |
| 4 | rs11722288 | TACR3 | 0.121 | 0.148 | *0.061 | 0.266 |
| 7 | rs10499934 | ACN9 | 0.859 | 0.385 | 0.224 | 0.412 |
| 7 | rs7794886 | ACN9 | 0.941 | 0.476 | 0.590 | 0.512 |
| 7 | rs12671685 | ACN9 | 0.746 | 0.452 | 0.252 | 0.307 |
| 7 | rs324649 | CHRM2 | 0.868 | 0.610 | 0.429 | 0.121 |
| 8 | rs997917 | OPRK1 | 0.937 | 0.989 | 0.956 | 0.954 |
| 8 | rs16918941 | OPRK1 | 0.516 | 0.712 | 0.773 | 0.499 |
| 8 | rs6985606 | OPRK1 | 0.495 | 0.439 | 0.851 | 0.522 |
| 11 | rs17115439 | ANKK1 | *0.077 | 0.238 | 0.964 | 0.825 |
| 11 | rs1799978 | DRD2 | 0.133 | **0.040 | 0.239 | 0.480 |
| 15 | rs1897356 | GABRB3 | 0.570 | 0.847 | *0.064 | **0.048 |
| 15 | rs1571281 | GABRG3 | 0.296 | 0.749 | 0.926 | 0.905 |
| 15 | rs680244 | CHRNA5 | 0.779 | 0.923 | 0.909 | 0.239 |
| 20 | rs2235749 | PDYN | 0.696 | 0.680 | 0.513 | 0.381 |
| 20 | rs6045819 | PDYN | 0.840 | 0.687 | 0.652 | 0.535 |
| 20 | rs1997794 | PDYN | 0.255 | 0.655 | 0.470 | 0.833 |
- P values are shown for logistic regression results of each individual SNP for association with AD. ‘P-val COGA EA’ indicates results of association analyses in the European American subset of the COGA GWAS sample that is independent of the COGA high-density family-based association sample. ‘P-val SAGE EA’ reflects association results in the FSCD and COGEND portion of the SAGE European American sample. ‘COGA All’ and ‘SAGE All’ show results in samples that are included in the EA portion of the COGA high-density family-based association sample, as well as independent individuals of other ancestries. **SNPs with P < 0.05 for association with AD, *SNPs with P < 0.10. COGA = Collaborative Study on the Genetics of Alcoholism; GWAS = genome-wide association studies; SAGE = Study of Addiction: Genes and Environment; SNP = single nucleotide polymorphism.
ROC curve analysis
The distribution of genetic sum scores was similar in cases and controls in COGA and SAGE (Fig. 2). Neither of the genetic sum scores had an AUC estimate that reached statistical significance at P < 0.05 for COGA or SAGE. Because of the lack of replication for individual SNPs and sum score associations with AD, AUC estimates were not significant. Family history, however, did produce a statistically significant AUC. ROC curve analysis results for family history compared with the sum scores are summarized in Table 5.
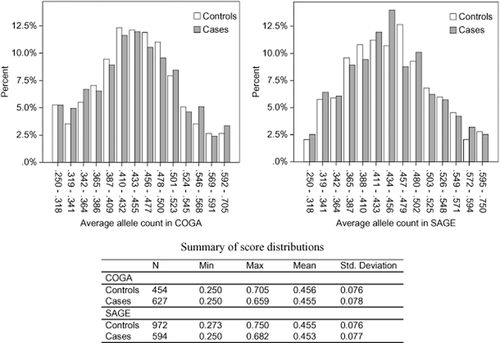
Distribution of genetic sum scores based on candidate gene single nucleotide polymorphisms (SNPs) pruned at r2 < 0.50 in cases and controls for alcohol dependence (AD). Left panel: scores in the Collaborative Study on the Genetics of Alcoholism (COGA) genome-wide association study (GWAS) sample independent of the COGA high-density family-based association sample. Right panel: scores in the Family Study of Cocaine Dependence and Collaborative Genetic Study of Nicotine Dependence portion of the Study of Addiction: Genes and Environment (SAGE) GWAS sample. The figure shows the frequency of normalized allele counts in bins separately for cases and controls. Allele counts were created by adding the number of risk alleles of SNPs associated with AD in candidate gene studies, and then dividing by the number of non-missing genotypes for each individual. The table summarizes the mean and range for the sum score in cases and controls
| Diagnostic classifier | AUC | Standard errora | Asymptotic significanceb | Asymptotic 95% confidence interval | |
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| COGA | |||||
| Family historyd | 0.686 | 0.016 | <0.001 | 0.654 | 0.718 |
| Genetic sum scorec | 0.498 | 0.018 | 0.915 | 0.463 | 0.533 |
| SAGE | |||||
| Family historyd | 0.614 | 0.015 | <0.001 | 0.584 | 0.643 |
| Genetic sum scorec | 0.496 | 0.015 | 0.782 | 0.466 | 0.525 |
- aUnder the non-parametric assumption. bNull hypothesis: true area = 0.5. cGenetic sum score based on pruned list of COGA variants at an r2 of 0.50. dFamily history was determined by a binary absence or presence of family history of AD in COGA and the presence or absence of parental AD in SAGE. AUC = area under the receiver operating characteristic curve; COGA = Collaborative Study on the Genetics of Alcoholism; GWAS = genome-wide association studies; SAGE = Study of Addiction: Genes and Environment.
GWAS results from varying P-value thresholds
Table 6 summarizes mean AUC estimates and median P values for each set of SNPs meeting P-value thresholds across the 100 random divisions of the SAGE–COGA combined sample. AUC estimates were significant at P < 0.05 for subsets of SNPs meeting P-value thresholds of 0.01 and greater. Figure 3 illustrates the AUC estimates of genetic sum scores created based on varying P-value thresholds. Although the P-value threshold at which AUC value peaked varied across subsets, AUC point estimates showed an increasing trend across the subsets as the P-value threshold used for SNP selection became less stringent.
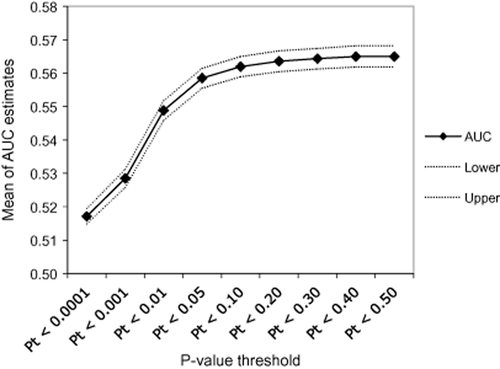
Mean area under the receiver operating characteristic curve (AUC) estimates for varying P-value thresholds. The mean of all 100 AUC estimates for sum scores created using single nucleotide polymorphisms that meet different P-value thresholds in discovery samples is plotted here in solid line. Dashed lines represent the upper and lower bounds of the 95% confidence interval of the mean AUC estimate
| P-value threshold | Mean AUC | 95% confidence interval | Median P value for AUC | |
|---|---|---|---|---|
| Lower | Upper | |||
| Pt < 0.50 | 0.565 | 0.562 | 0.568 | 1.37E-05 |
| Pt < 0.40 | 0.565 | 0.562 | 0.568 | 1.42E-05 |
| Pt < 0.30 | 0.564 | 0.561 | 0.567 | 1.82E-05 |
| Pt < 0.20 | 0.564 | 0.561 | 0.567 | 2.62E-05 |
| Pt < 0.10 | 0.562 | 0.559 | 0.565 | 4.81E-05 |
| Pt < 0.05 | 0.559 | 0.556 | 0.562 | 1.04E-04 |
| Pt < 0.01 | 0.549 | 0.546 | 0.552 | 0.00166 |
| Pt < 0.001 | 0.528 | 0.526 | 0.531 | 0.0631 |
| Pt < 0.0001 | 0.517 | 0.515 | 0.519 | 0.29 |
- Summary statistics for 100 random 50% splits of the combined COGA–SAGE sample into discovery samples and validation samples. Sum scores were created based on SNPs meeting each P-value threshold by adding minor alleles weighted by the log of the odds ratio for AD. Confidence intervals are based on 100 AUC estimates from 100 separate sum score calculations at each P-value threshold. Median P-value threshold was calculated because distributions of P values were skewed. AUC = area under the receiver operating characteristic curve; SNP = single nucleotide polymorphism.
Discussion
This study aimed to evaluate the clinical validity of genetic variants that have been associated with AD by exploring the aggregate effect of associated SNPs on risk prediction for AD. Prior studies on the clinical use of genetic information in predicting risk for other complex disorders have investigated the effect of genetic sum scores in risk assessment and shown significant, but small, AUCs. In our study, genetic sum scores were created based on results from two different sources: SNPs that were associated with AD in family-based candidate gene studies and SNPs from GWAS analyses that met varying P-value thresholds. ROC curve analysis was used to assess the ability of the sum scores to classify cases and controls for AD.
Results did not show significant AUCs for the candidate gene sum scores, suggesting that sum scores of this limited set of SNPs are not predicting better than chance. The individual variants contributing to the sum scores did not yield significant results in the independent samples in which discriminative ability was assessed. Results from the GWAS analyses resulted in significant, albeit small, AUC estimates for P-value thresholds of 0.01–0.50. These results support a polygenic model involving hundreds of variants of small effect contributing to risk for AD that is consistent with previous findings on schizophrenia and bipolar disorder (Purcell et al. 2009). Less-stringent thresholds allowed for the selection of more true findings with effect sizes that would not otherwise have reached genome-wide significance. Combining nominally associated SNPs in aggregate improved clinical validity because these true loci could outweigh noise from null loci.
This assessment of discriminatory accuracy shows that these panels of SNPs currently have limited clinical utility. One reason that many of the candidate gene SNPs did not replicate in the independent samples used to assess for clinical validity could be due to heterogeneity across samples; different genetic variants may contribute to risk in different populations containing varying subsets of alcohol-dependent individuals. Therefore, genetic risk could be unique to the samples used in these association analyses. For example, several variants have been found to have stronger association with AD in individuals with co-occurring drug dependence. Dick et al. showed that CHRM2 is associated with a form of AD that is co-morbid with drug dependence, but not with AD alone (Dick et al. 2007a). In another case, Foroud et al. found that SNPs in TACR3 that were associated with AD in EA COGA families had the strongest association in individuals with more severe AD and co-morbid cocaine dependence (Foroud et al. 2008). Furthermore, Agrawal et al. showed that GABRA2 is associated with AD only in individuals with co-morbid drug dependence. When these individuals were removed from the analysis, no association remained (Agrawal et al. 2006). A future step in developing genetic risk models for AD would be to assess for prediction for different subtypes of AD.
SNPs from primary analyses in the family-based portion of the study may not have replicated in independent COGA and SAGE GWAS individuals due to sampling differences between the GWAS samples and the family-based association sample. One possibility is that the high-density family-based sample may be more severely affected than a case-control sample and therefore show differences in underlying genetic etiology. Mean DSM-IV symptom counts for AD were similar across the COGA high-density family-based sample (mean = 5.26, SD = 1.48), and the SAGE (mean = 4.87, SD = 1.51) and COGA GWAS samples (mean = 5.56, SD = 1.43); however, severity of AD may differ in ways beyond criterion count, such as the severity of the symptoms themselves, including the extent of tolerance and withdrawal, duration of symptoms, and number of episodes. We combined the COGA and SAGE samples before performing subsampling in order to create samples with similar population structure across discovery and validation sets.
We also created discovery and replication samples by splitting just the FSCD and COGEND portion of the SAGE GWAS sample in half, and then assessing for clinical validity in the COGA GWAS sample. Of the list of SNPs that met nominal significance criteria in both halves of the SAGE sample, the majority of SNPs did not share the same direction of effect, suggesting that many of these results could be false positives. This study also explored the effect of using a more stringent r2 threshold of 0.25 to prune the list of candidate gene SNPs before creating sum scores; results were similar.
These results show that family history is a better classifier than current conceptualizations of SNP panels, based on candidate gene and GWAS for AD. Family history is likely a better predictor than this panel of SNPs because it accounts for more of the latent genetic factors contributing to AD, whereas the contribution to risk of the panel of SNPs is less clear. Family history also contains non-genetic predictors, which could account for a significant proportion of the risk as well, as family history could influence to some extent the environment that an individual is exposed to during development. Furthermore, the etiology of AD may be different for one family versus another. Therefore, risk prediction based on an individual's family history may encompass genetic factors that are more specific to that individual than a general panel of SNPs, which may not explain risk for the particular subgroup to which that individual belongs. We assessed the value of combining information from the candidate gene panel with family history, as family history and the candidate gene sum score were not correlated (r = 0.021, n = 1081, P = 0.490). We found that the AUC for family history increased nominally from 0.686 to 0.690 in COGA after adding the candidate gene sum score. This suggests that there was negligible additional information when the candidate gene panel was added to family history information.
Importantly, before assessment of clinical validity is made, the contribution of genetic sum scores, rather than individual associated SNPs, must be determined. The finding that genetic sum scores created from SNPs meeting less-stringent P-value thresholds were significantly associated with AD and had significant discriminative ability suggests that varying P-value thresholds could better detect variants of small effect. However, it is difficult to distinguish true alleles of vanishingly small effect from alleles in LD with causal alleles. Because variants contributing to AD have small effect sizes, and the outcome used in the association studies is a dichotomous diagnosis rather than a continuous outcome, larger sample sizes are needed for increased power to detect causal variants that replicate across studies (Bierut et al. 2010). The samples used in this study did not have enough power to detect the entire range of small effect sizes for individual variants assessed in these analyses at a genome-wide significance level. Splitting the COGA–SAGE combined sample further reduced power.
GWA studies have shown replication of SNPs associated with AD in the COGA candidate gene studies (Bierut et al. 2010; Edenberg et al. 2010); however, in an effort to create SNPs that captured unique information by pruning them based on LD, some of the replicated SNPs were not included in the model. An expanded candidate gene sum score incorporated more SNPs that met nominal significance levels in the COGA and SAGE GWAS samples (Supporting Information Table S1), but did not have a significantly different AUC compared with the candidate gene sum score composed of pruned SNPs. In these data, we have previously demonstrated that the missense SNP rs1229984 is associated with AD at P < 5 × 10−8 (Bierut et al. 2012). This variant, previously well recognized for its protective influence on alcoholism in Asians, has also been found to exert an influence on alcoholism risk in Caucasians and AAs. However, it is fairly uncommon in non-Asian samples (<5%) and is poorly captured by content on commercially available GWAS platforms due to lack of LD with neighboring SNPs. We assessed the discriminatory accuracy of this ADH1B SNP for AD and found that it alone had an AUC of 0.538 (P = 7.58 × 10−4) in COGA. The inclusion of this SNP in the candidate gene sum score increased the AUC from 0.498 to 0.503, but this AUC was not significant (P = 0.885), presumably due to the very low-allele frequency in this population. This suggests that including known variants that replicate in the validation sample used for prediction could have a greater AUC. Expanding the panel to include additional replicated variants could increase the AUC further.
A prediction model that consists primarily of genetic variants has a maximum AUC constrained by the heritability of the trait, as well as the disease prevalence in a population (Wray et al. 2010). As heritability of a disease goes down and as prevalence goes up, the maximum AUC goes down (Wray et al. 2010). This stresses the importance of taking into account other factors contributing to the variability in AD for risk prediction, particularly because AD is a fairly prevalent disorder. Additional measures to increase power may include reducing heterogeneity by refining the phenotype used as the outcome in the association study (Bierut et al. 2010). Large-scale meta-analysis, along with expanded individual association studies for AD, may improve the detection of disease variants.
We do not yet have enough information about the specific variants contributing to AD to use genetic data for clinical risk prediction. These findings conclude that despite interest in genetic testing, and availability of testing through DTC avenues, genetic testing for AD is not yet ready to be applied in a clinical setting. This study suggests that expanding the number of replicated variants associated with AD would account for a greater portion of the genetic variance for AD and therefore improve risk prediction. Because AD also has a substantial unique environmental etiology in addition to genetics, a prediction tool based on genetic information alone would not have the highest AUC; the addition of environmental factors that would account for more of the variability in AD and therefore a model that takes into consideration both could have better predictive ability. Data simulations in our study show that adding environmental effects could potentially raise the predictive accuracy to 0.95 (Maher et al., in preparation). While genetic information may be of limited clinical validity at the moment, as we continue to identify genes successfully, and incorporate information from both genetic and environmental risk factors, there is potential for future clinical utility.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B.P., V. Hesselbrock, H.E., L. Bierut, includes 10 different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J.E., J.N. Jr., T. Foroud); University of Iowa (S.K., J.K.); SUNY Downstate (B.P.); Washington University in St. Louis (L. Bierut, A.G., J.R., K. Bucholz); University of California at San Diego (M.S.); Rutgers University (J.T.); Southwest Foundation (L.A.), Howard University (R. Taylor); and Virginia Commonwealth University (D.D.). Other COGA collaborators include L. Bauer (University of Connecticut); D. Koller, S. O'Connor, L. Wetherill, X. Xuei (Indiana University); Grace Chan (University of Iowa); N. Manz, M. Rangaswamy (SUNY Downstate); A. Hinrichs, J. Rohrbaugh, J-C. Wang (Washington University in St. Louis); A. Brooks (Rutgers University); and F.A. (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). This work was also funded by K02AA018755 (D.M.D).
The COGA GWAS sample genotyping was funded through U10 AA008401 from the NIAAA, National Institutes of Health, Bethesda, MD, USA, U01HG004438, and NIH GEI, and HHSN268200782096C, NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’. The genotyping center was the Johns Hopkins University Center for Inherited Disease Research (CIDR), Baltimore, MD, USA.
SAGE is part of the Gene Environment Association Studies initiative (GENEVA) funded by the National Human Genome Research Institute. Funding sources are: U01 HG004422, National Human Genome Research Institute, Bethesda, MD, USA; U10 AA008401, National Institute on Alcohol Abuse and Alcoholism and National Institute on Drug Abuse, Bethesda, MD, USA; P01 CA089392, National Cancer Institute, Bethesda, MD, USA; R01 DA013423, National Institute on Drug Abuse, Bethesda, MD, USA; and R01 DA019963, National Institute on Drug Abuse, Bethesda, MD, USA.
Funding sources did not have a role in the design, conduct or reporting of this study.
Disclosure/Conflict of Interest
JPR and AMG are inventors on the patent ‘Markers for Addiction’ (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction.
Authors Contribution
JY and DMD drafted the manuscript and were responsible for the primary study concept, design and interpretation of findings. JY was responsible for the primary statistical analyses and draft of the manuscript. FA made key contributions to data analysis and data management. BTW and KSK critically reviewed drafts of the manuscript and provided insights into the direction and interpretation of the analyses. VSW contributed to the determination of proxy SNP information. HJE, AA, LA, JIN, MAS, JRK, JPR, SK, AMG, JAT and BP contributed to data collection and management and review of the manuscript. HJE, AA, MKZ, LA, JIN, MAS, JRK, JPR and SK provided valuable feedback on the analyses and interpretation of results.




