Dysbiosis in the Gut Microbiome of Pembrolizumab-Treated Non-Small Lung Cancer Patients Compared to Healthy Controls Characterized Through Opportunistic Sampling
Funding: This work was supported in part by a research grant from the Investigator-Initiated Studies Program of Merck Sharp & Dohme Corp.
ABSTRACT
Background
The gut microbiome influences the host immune system, cancer development and progression, as well as the response to immunotherapy during cancer treatment. Here, we analyse the composition of the gut bacteriome in metastatic Non-Small Cell Lung Cancer (NSCLC) patients receiving Pembrolizumab immunotherapy within a prospective maintenance trial through opportunistic sampling during treatment.
Methods
The gut microbiome profiles of NSCLC patients were obtained from stool samples collected during Pembrolizumab treatment and analysed with 16S rRNA metagenomics sequencing. Patient profiles were compared to a group of healthy individuals of matching ethnic group, age, sex, BMI and comorbidities.
Results
A significant decrease in the treated patients was observed in two prominent bacterial families of the phylum Firmicutes, Lachnospiraceae and Ruminoccocaceae, which comprised 31.6% and 21.8% of the bacteriome in the healthy group but only 10.9% and 14.2% in the treated patient group, respectively. Species within the Lachnospiraceae and Ruminococcaceae families are known to break down undigested carbohydrates generating short chain fatty acids (SCFA), such as butyrate, acetate and propionate as their major fermentation end-products, which have been implicated in modifying host immune responses. In addition, a significant increase of the Bacteroidacaeae family (Bacteroidetes phylum) was observed from 10.7% in the healthy group to 23.3% in the treated patient group. Moreover, and in agreement with previous studies, a decrease in the Firmicutes to Bacteroidetes ratio in the metastatic NSCLC Pembrolizumab-treated patients was observed.
Conclusion
The observed differences indicate dysbiosis and a compromised intestinal health status in the metastatic NSCLC Pembrolizumab-treated patients. This data could inform future studies of immunotherapy treatment responses and modulation of the gut microbiome to minimise dysbiosis prior or concurrent to treatment.
Trial Registration: SWIPE Trial (NCT 02705820).
1 Introduction
Lung Cancer (LC) is the leading cause of cancer mortality worldwide, and the most recent data derived from Global Cancer Observatory 2022 has shown a significant increase in both global incidence and mortality of LC [1]. Over 2.4 million new LC cases and more than 1.8 million deaths were recorded in 2022. There are two main subdivisions of lung cancer: Small Cell Lung Cancer (SCLC) and Non-Small Cell Lung Cancer (NSCLC), which account for about 85% of all patients with LC. The main causative factor to develop LC remains tobacco smoking, while other environmental (asbestos, radon, environmental pollution) and genetic predisposition factors for some individuals [2] also play a role. Recent studies have demonstrated that the interaction of the intestinal microbiota and the host's immune system can promote carcinogenesis [3].
The human microbiota of the human body, including those of the skin, gastrointestinal tract, genitourinary system, and respiratory tract, form communities of bacteria, archaea, fungi, viruses, protists, and eukaryotic microorganisms. They participate in key functions of the human body, including metabolic, nutritional, physiological, and immunological processes [4], as well as susceptibility to cancer, through their immense metabolic capacity and influence on immunity [5]. The development of next-generation sequencing technologies, specifically the sequencing of the 16S rRNA gene, has made it possible to accurately characterize the entire gut bacteriome of patients in an accessible and culture-free manner. Gut microbiome studies have revealed significant differences in the relative abundance of certain microbes in patients with cancer compared with healthy controls [6]. Association studies, however, cannot differentiate between cause and effect [5]. Preclinical studies using mouse models for oncogenesis shed more light in this debate and suggest a causative role, in that microbiota can influence cancer susceptibility and progression by diverse mechanisms such as by modulating inflammation, influencing the genomic stability of host cells, and producing metabolites that function as histone deacetylase inhibitors to epigenetically regulate host gene expression [5, 6].
More recently, great progress has been achieved in the treatment of metastatic NSCLC with the use of Immune Checkpoint Inhibitors (ICIs). ICIs target T-cell regulatory pathways, specifically against the programmed cell death protein-1 (PD-1) and against the Programmed Death Ligand 1 (PD-L1), as well as the Cytotoxic T-Lymphocyte-Associated protein 4 (CTLA-4). Over the last few years, durable responses with ICIs in patients with metastatic NSCLC, as well as other malignancies, have been observed, with clinical studies showing significant survival benefit, leading to the approval of these agents by regulatory authorities and their use in standard clinical practice [7]. However, effective responses to ICIs are reported in a minority of cancer patients, mainly due to limited immune cell infiltration into tumors or due to an immunosuppressive tumor microenvironment [8], and there is an urgent need to identify reliable predictive biomarkers for immunotherapy efficacy.
A number of preclinical and animal studies have shown the influence of the gut bacteria on the antitumor efficacy of both chemotherapy [9, 10] and immunotherapy agents [11, 12], suggesting that bacteria-mediated interactions with the immune system are essential for optimal drug efficacy [8]. Furthermore, three landmark studies in 2018 confirmed these findings in patients receiving immunotherapy for advanced metastatic cancer, showing that patients can be stratified into responders and non-responders to immunotherapy on the basis of the composition of their intestinal microbiomes [13-15], using 16S rRNA sequencing, provoking great interest in this field.
In this pilot study based on a small number of patients, we sought to characterize dysbiosis in the gut microbiome of metastatic NSCLC patients receiving pembrolizumab immunotherapy in a prospective maintenance study through opportunistic sampling of patients during their treatment. We compared the fecal bacteria between NSCLC patients and a group of healthy individuals of matching age, sex, BMI, comorbidities, and ethnic background in order to characterize the changes in microbial populations resulting from dysbiosis typically experienced by these patients. Our secondary objective was to investigate the functional impact of the abundance of short-chain fatty acid (SCFA) bacteria and determine if the gut microbiome could be associated with response to pembrolizumab immunotherapy. Without pre-treatment/baseline samples prior to chemotherapy and immunotherapy, the differences observed in the gut microbiome may be due to the underlying disease, that is, the metastatic NSCLC, the previous platinum-based chemotherapy, and pembrolizumab immunotherapy that all patients received, or due to the progression on treatment; therefore, we can only conclude broad differences in bacterial profiles between healthy individuals and NSCLC patients. However, the results provide insight into the degree of dysbiosis present in metastatic NSCLC patients who have received chemotherapy and immunotherapy that needs to be interpreted in conjunction with other evidence regarding dysbiosis and immunotherapy efficacy.
2 Methods
2.1 Study Groups
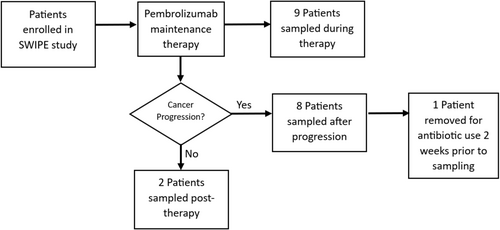
Stool samples were obtained opportunistically from nineteen NSCLC patients enrolled in a prospective study of maintenance treatment using pembrolizumab (with informed consent and stool sampling protocol approval ЕЕВК/ЕП/2016/15 from the Cyprus National Bioethics Committee). One patient was excluded due to having received antibiotics two weeks prior to the stool sample collection; hence, eighteen patients were available for analysis. Nine samples were collected during pembrolizumab immunotherapy, two within the post-immunotherapy surveillance period (after 24 months of pembrolizumab treatment), and seven after starting chemotherapy due to cancer progression on pembrolizumab immunotherapy (Figure 1). While no pre-treatment control samples were obtained, stool samples from a control group in a concurrent clinical study for colorectal cancer screening consisted of 154 healthy volunteers who were matched for age, sex, BMI, comorbidities (cardiovascular disease and diabetes) and ethnic background to the study population and were sequenced at the same time with the same methods [16]. Patient details are given in Table 1.
| Sex | NSCLC treated | Healthy controls | ||
|---|---|---|---|---|
| Number | Mean age | Number | Mean age | |
| Male | 13 | 62.23 | 87 | 63.36 |
| Female | 5 | 66.20 | 67 | 65.86 |
| All | 18 | 63.33 | 154 | 64.45 |
Finally, an exploratory analysis was undertaken due to the small sample size to investigate differences in the microbiome of metastatic NSCLC patients on pembrolizumab immunotherapy with durable clinical benefit (defined as duration on treatment > 6 months) versus those without durable clinical benefit.
2.2 16S-Sequencing
Fecal bacteria DNA isolation and purification were performed with either the PureLink Microbiome DNA Purification Kit (Invitrogen) or ZYMO microbiome kit (Zymo Research Corporation, USA) using approximately 0.2 g from the initial fecal sample. After extraction, 90 μL of eluted DNA was stored at −80°C until analysis. The microbiota was characterized using the V3–V4 hypervariable regions of the 16S rRNA gene using standard primers [17]. These were sequenced using the Nextera XT Library Preparation kit (Illumina Inc., San Diego, CA, United States) [18], with a 300 bp paired-end run on an Illumina MiSeq platform, following the standard Illumina protocols.
2.3 Bioinformatics and Statistical Analysis
Bioinformatics analysis, including sequence quality and filtering, was based on the 16S Metagenomics app (v1.1.0) of the Illumina Basespace platform. Operational taxonomy unit (OTU) classification analyses were performed using the Ribosomal Database Project Classifier [19] against the Illumina-curated version of the Genome Taxonomy Database (GTDB) (version 4.3, available at: doi: 10.5281/zenodo.6655692). This resulted in the identification and calculation of the relative abundance of each bacterium for taxonomy levels from family to species. The data was normalized by the average number of reads per sample. Quality control was applied using a minimum detection of a bacterium in at least 30% of samples and a minimum average relative abundance of over 1% in at least one group.
The Mann–Whitney U test was employed to compare the bacterial abundance between groups for each taxonomy level (Family, Genus, and Species). This non-parametric test was chosen due to the small sample size of the study and its ability to assess differences even when the data is not normally distributed. To further validate the robustness of the observed differences, permutation testing using the EnvStats R package [20], with 10 000 000 permutations, was conducted on the statistically significant findings identified using the Mann–Whitney U test. This resampling-based method evaluates the significance of observed differences by randomly rearranging the data while preserving the overall group structure. Permutation testing plays a crucial role in small sample size studies, as it helps to eliminate false positives caused by random fluctuations and provides stronger evidence for the true existence of significant differences. The combination of Mann–Whitney U tests and permutation testing provides a comprehensive approach to assessing and validating the observed differences in bacterial abundance between the study groups. This rigorous methodology ensures that the conclusions drawn from the study are firmly grounded in the data and robust to the limitations of small sample size [21, 22]. Alpha diversity was calculated using the Simpson Index (weighted mean of the proportional abundances). Microbial community analysis was performed on 16S rRNA sequencing data using PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States), a software for predicting functional abundances based only on marker gene sequences [23]. This analysis facilitated the identification and quantification of specific enzymes (Enzyme Commission numbers) and metabolic pathways (KEGG orthologs).
2.4 Sequencing Depth
Mean sequencing depth (summarized in the Supporting Information) was high for all samples, with a mean of approximately 90 000 reads across all samples. The minimum number of reads for a sample was 23 000 reads in the control group. This ensured that OTU assignment could be achieved at high confidence, reaching over 94% mean classification to genus in both groups.
3 Results
3.1 Bacterial Diversity
Mean alpha diversity statistics are shown in Table 2 and Figure 2. The control group indicated a more diverse bacterial community than treated NSCLC patients at the genus (p < 2.14E-06) and species (p < 0.00011) levels. At the family level, there was no significant difference between the two groups (p < 0.078).
| Level | Mean alpha diversity | ||
|---|---|---|---|
| NSCLC treated (n = 18) | Healthy controls (n = 154) | p | |
| Family | 5.324 | 4.834 | 0.078 |
| Genus | 7.205 | 12.442 | 2.14E-06 |
| Species | 10.856 | 16.308 | 0.00011 |
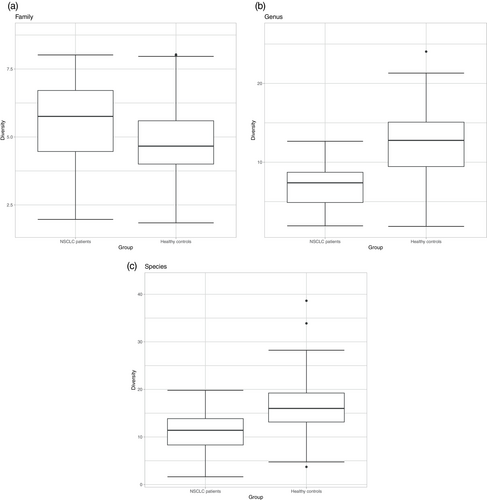
3.2 Relative Abundance
A summary of the relative abundance analysis at each taxonomic level (phylum, family, genus, species) is shown in Tables 3–6, respectively. These tables indicate the bacteria that were above the 1% relative abundance threshold in one or both groups and found to be significant at the p < 0.01 level using a permutation test of mean relative abundance between groups.
| Phylum | Mean relative abundance | ||
|---|---|---|---|
| NSCLC treated (n = 18) | Healthy controls (n = 154) | p | |
| Actinobacteria | 2.573 | 8.18 | 1.87E-05 |
| Bacteroidetes | 33.71 | 16.78 | 1.33E-05 |
| Firmicutes | 45.89 | 70.73 | 5.48E-08 |
| Fusobacteria | 1.16 | 0.04 | 0.00215 |
| Proteobacteria | 7.92 | 2.34 | 5.46E-06 |
| Family | Mean relative abundance | ||
|---|---|---|---|
| NSCLC treated (n = 18) | Healthy controls (n = 154) | p | |
| Acidaminococcaceae | 5.038 | 1.440 | 1.00E-07 |
| Bacteroidaceae | 23.325 | 10.743 | 0.0002 |
| Coriobacteriaceae | 0.831 | 3.430 | 0.0085 |
| Erysipelotrichaceae | 0.468 | 2.552 | 0.0082 |
| Fusobacteriaceae | 1.164 | 0.037 | 1.00E-07 |
| Lachnospiraceae | 10.903 | 31.620 | 1.00E-07 |
| Porphyromonadaceae | 4.292 | 1.945 | 1.00E-07 |
| Rikenellaceae | 4.789 | 0.922 | 1.00E-07 |
| Ruminococcaceae | 14.198 | 21.815 | 0.0007 |
| Genus | Mean relative abundance | ||
|---|---|---|---|
| NSCLC treated (n = 18) | Healthy controls (n = 154) | p | |
| Acidaminococcus | 2.598 | 0.360 | 0.0001 |
| Alistipes | 4.898 | 0.951 | 1.00E-07 |
| Bacteroides | 23.809 | 11.110 | 0.0002 |
| Blautia | 1.140 | 8.737 | 1.00E-07 |
| Coprococcus | 0.390 | 2.371 | 0.008 |
| Dorea | 0.619 | 2.123 | 0.001 |
| Escherichia/Shigella | 3.463 | 1.260 | 0.0087 |
| Faecalibacterium | 2.464 | 8.324 | 1.00E-07 |
| Fusicatenibacter | 0.736 | 1.698 | 0.0047 |
| Lachnospiracea_incertae_sedis | 2.040 | 5.234 | 1.00E-07 |
| Oscillibacter | 1.731 | 0.681 | 0.0001 |
| Parabacteroides | 1.927 | 0.823 | 0.0001 |
| Ruminococcus | 2.027 | 6.384 | 0.0018 |
| Ruminococcus2 | 0.815 | 2.332 | 0.0029 |
| Species | Mean relative abundance | ||
|---|---|---|---|
| NSCLC treated (n = 18) | Healthy controls (n = 154) | p | |
| Acidaminococcus intestini (AF473835) | 2.141 | 0.242 | 0.0006 |
| Alistipes onderdonkii (AY974071) | 2.354 | 0.316 | 1.00E-07 |
| Alistipes senegalensis (NR 118219.1) | 1.422 | 0.349 | 1.00E-07 |
| Bacteroides_fragilis (CR626927) | 1.661 | 0.275 | 0.0011 |
| Bacteroides uniformis (AB050110) | 5.856 | 2.129 | 1.00E-07 |
| Blautia_luti (AJ133124) | 0.190 | 2.227 | 0.0033 |
| Blautia wexlerae (EF036467) | 0.562 | 3.415 | 0.0011 |
| Coprococcus comes (EF031542) | 0.227 | 1.109 | 0.0021 |
| Dorea longicatena (AJ132842) | 0.642 | 2.059 | 0.001 |
| Faecalibacterium prausnitzii (AJ413954) | 2.860 | 9.775 | 1.00E-07 |
| Fusicatenibacter_saccharivorans (AB698910) | 0.854 | 2.00 | 0.013 |
| Eubacterium hallii (L34621) | 0.287 | 2.106 | 0.0001 |
| Roseburia_faecis (AY305310) | 0.441 | 2.082 | 0.0099 |
| Veillonella dispar (AF439639) | 1.143 | 0.424 | 0.0001 |
At the phylum level (Table 3), Bacteroidetes were significantly more abundant in the NSCLC patients (33.7%) compared to the control group (16.7%). In contrast, Firmicutes were markedly decreased in the NSCLC patients (45.8%) compared to the control group (70.7%). Of the less abundant phyla detected, Actinobacteria was more abundant in the control group (8.18%) compared to 2.5% in the NSCLC patients, whereas Fusobacteria and Proteobacteria were increased in NSCLC patients (1.15% and 7.9%, respectively) compared to controls (0.04% and 2.3%, respectively).
At the family level (Table 4), ten bacteria families were noted to have a significant difference in mean relative abundance in the two groups. Acidaminococcaceae, Bacteroidaceae, Enterobacteriaceae, Fusobacteriaceae, Porphyromonadaceae, and Rikenellaceae were increased in treated NSCLC patients, whereas Coriobacteriaceae, Erysipelotrichaceae, Lachnospiraceae, and Ruminococcaceae were more abundant in the control group.
At the genus level (Table 5), fourteen bacteria genera were significantly different between the two groups. In the NSCLC group, Acidaminococcus, Alistipes, Bacteroides, Escherichia/Shigella, Oscillibacter, and Parabacteroides were increased. In the control group, Blautia, Coprococcus, Dorea, Faecalibacterium, Fusicatenibacter, Lachnospiracea_incertae_sedis, Ruminococcus, and Ruminococcus2 were more abundant.
At the species level (Table 6), fourteen bacteria were significantly different between the two groups. In the NSCLC group, Acidaminococcus intestini, Alistipes onderdonkii, Alistipes senegalensis, Bacteroides uniformis, Bacteroides fragilis, and Veillonella dispar were more abundant. In the control group, Blautia luti, Blautia wexlerae, Coprococcus comes, Dorea longicatena, Eubacterium hallii, Faecalibacterium prausnitzii, Fusicatenibacter_saccharivorans, and Roseburia_faecis were more abundant.
Full results of the Mann–Whitney U tests of mean relative abundance are presented in the Supporting Information.
3.3 Functional Impact of the Butyrate Pathway
A summary of the functional analysis of potential butyrate metabolic pathways associated with the microbiome species composition is shown in Figure 3. The figure indicates enzymes within two of the key metabolic pathways of butyrate production—acetyl-CoA and lysine—showed significant differences between the two groups.
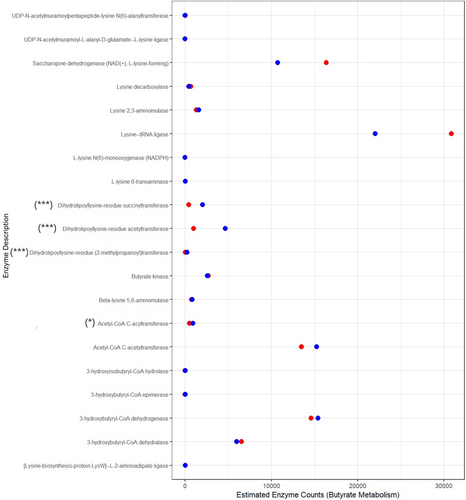
 NSCLC patients
NSCLC patients  Healthy controls) significance of difference between the means based on Wilcoxon signed rank test (***p < 0.001, **p < 0.01, *p < 0.05).
Healthy controls) significance of difference between the means based on Wilcoxon signed rank test (***p < 0.001, **p < 0.01, *p < 0.05).3.4 Microbiome and Immunotherapy Efficacy
No statistically significant differences in the bacteriome were observed between patients with durable clinical benefit (> 6 months on pembrolizumab treatment) and no clinical benefit to treatment in relation to bacteria with greater than 1% relative abundance.
4 Discussion
In this study, we reveal a significant decrease in mean alpha diversity at two taxonomic ranks, genus and species, in the NSCLC patient group treated with ICIs compared to a healthy control group. Gut dysbiosis and lower bacterial diversity are common in cancer patients, leading to a compromised immune system [24] whereas greater bacterial diversity has been linked to both overall survival [25, 26] and progression-free survival [27, 28]. Additionally, our study revealed a lower ratio of Firmicutes/Bacteroidetes (F/B) in NSCLC patients versus the control group, a finding similar to that in previous studies comparing the gut microbiome of NSCLC patients treated with ICIs with healthy individuals [28-32]. The F/B ratio is thought to have an important influence in maintaining normal intestinal homeostasis. An altered F/B ratio is regarded as dysbiosis, whereby an increased F/B ratio is usually observed with obesity, and a decreased F/B ratio with conditions such as inflammatory bowel disease (IBD) [33]. For example, low richness of the gut microbiome composition along with a reduced F/B ratio [34] may lead to low-grade chronic inflammation that stimulates T-cell exhaustion and decreases cytotoxic T cells [35, 36]. The significance of the lower F/B ratio relates to the fact that this leads to a lower concentration of SCFAs, which influence systemic immunity and inflammation [37-40]. SCFAs modify both cytokine production and dendritic cell activity, improving immune responses [41]. Butyrate, especially, is a crucial fatty acid associated with anti-inflammatory action and induces differentiation of regulatory T cells, cellular proliferation, and apoptosis by activating signaling pathways [42-44]. Several SCFAs are linked to the immune system through their association with the promotion and differentiation of colonic regulatory T cells (Tregs) [43, 45], thereby influencing inflammatory responses [46] and tumor-suppressing activity [47]. High concentrations of fecal acetic acid, propionic acid, butyric acid, and valeric acid in a clinical trial of cancer patients receiving either nivolumab or pembrolizumab immunotherapy have been associated with longer progression-free survival [48].
Our results also show significant changes in the bacterial relative abundance between the two groups; at the taxonomic rank of family, Lachnospiraceae had a three-fold decrease (10.9%) in treated NSCLC patients compared to controls (31.8%). Additionally, Ruminococcaceae was also found in significantly lower levels in the treated NSCLC patients (14.2%) relative to the control group (21.8%). Lachnospiraceae, Porphyromonadaceae, and Ruminococcaceae families are SCFA producers, belonging to the Firmicutes phylum. Most Lachnospiraceae and Ruminococcaceae members have the potential to maintain gut health by breaking down undigested carbohydrates and generating the SCFAs, butyrate, acetate, and propionate, via fermentation. Differences in the Lachnospiraceae family, in particular, were significant across all taxonomic levels (Figure 4) reinforcing the evidence for this finding. This family of bacteria has been associated with favorable clinical outcomes in immunotherapy treatment [49] and several other diseases [50].
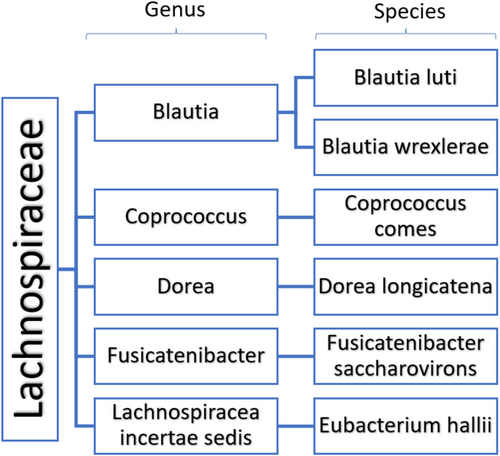
In our results, the SCFA producers Blautia luti, Coprococcus comes, Dorea, and Fusicatenibacter genera, and Ruminococcus spp. were all decreased in NSCLC patients, consistent with other studies [51, 52]. We also showed a significant increase in the Bacteroidaceae family in the treated NSCLC patient group (23.3%) relative to the control group (10.5%). This family involves bacteria with pro-inflammatory properties, like the prominent Bacteroides vulgatus. Multiple studies have linked commensal and opportunistic pathogenic bacteria to metabolic diseases. Elevated levels of Roseburia intestinalis, Eubacterium ventriosum, Ruminococcus torques, Ruminococcus gnavus, and Dorea longicatena have been associated with obesity [53], while B. vulgatus, Prevotella copris, and certain Clostridium spp., along with a decrease in the abundance of A. muciniphila and the loss of butyrate-generator taxa, correlate with type 2 diabetes mellitus [54].
Interestingly, the families Acidaminococcaceae, Bacteroidaceae, Enterobacteriaceae, and Rikenellaceae, also containing many pathogenic species, were more abundant in treated NSCLC patients. Higher abundance of Acidaminococcaceae family along with its members Acidaminococcus and Acidaminococcus intestini is in agreement with findings showing higher levels of Acidaminococcus in NSCLC patients with immune-related adverse effects upon anti-PD-1 antibody treatment [55]. The Enterobacteriaceae family includes common opportunistic pathogens, such as Escherichia, Shigella, and Klebsiella species, proposed to be “passenger bacteria” in colorectal cancer development [56, 57]. The Rikenellaceae family along with the Alistipes onderdonkii and Alistipes senegalensis species were found in higher abundance in treated NSCLC patients and require further investigation, since previous studies link their higher abundance in LC patients with antibiotic treatments prior to immunotherapy [58]. The increased prevalence of Rikenellaceae, Bacteroidaceae, and Enterobacteriaceae families in treated NSCLC patients has also been noted in previous studies [32, 59]. In addition, NSCLC patients showed lower abundance of Lachnospiraceae and Ruminococcaceae, which are all SCFA-producing bacteria that play a key role in metabolic interactions with the host and other microbes.
The gut bacteriome has a major role in regulating human health and disease, impacting metabolic, nutritional, physiological, and immunological processes in the human body, and helps to ensure homeostasis [4, 60]. Dysbiosis in the gut is associated with gastrointestinal tumors and IBD [61], but also with non-gastrointestinal conditions, such as obesity [62], cardiovascular disease [63] and diabetes [64]. Alterations in the gut bacteria composition, which impact intestinal homeostasis, are associated with major effects in the host's immunity and cancer initiation and progression [3, 65, 66], such that manipulation of the gut microbiome has also been suggested as a means of improving immunotherapeutic responses to ICIs [8, 13-15, 67]. The composition of the intestinal bacteriome is potentially a predictive indicator of the outcome of immunotherapy, which could allow stratification of patients into responders and non-responders to ICIs [68]. In particular, the presence of bacteria such as Akkermansia municiphilia [13], Faecalibacterium pausnitzii [14], Bifidobacterium longum [15], and others which share similar properties of butyrate and propionate production, has been found to influence sensitivity to ICIs and can mediate the immunotherapy effect [48]. This is due to immunomodulation of CD4+ cells, an increase in IFNγ and granzyme B in CD8+ cytotoxic T lymphocytes, and interleukin-17-secreting CD8+ T cells [69] which inhibits T-cell apoptosis, suppresses tumor growth, and upregulates antitumor immune responses [48]. In this study, perhaps due to the small sample size, we were unable to detect a difference in the bacteriome between patients with durable clinical benefit and no clinical benefit to pembrolizumab immunotherapy regimes.
Finally, predictive functional analysis of the microbiome profiles showed that the potential for butyrate metabolic pathways was different between the two groups in both the acetyl-CoA pathway and lysine pathways. The acetyl-CoA pathway is used by commensal bacteria to convert lactate and acetate into butyrate [70] whereas the lysine pathway is dominated by members of the Bacteroidetes [71]. However, functional analysis is only an estimate of potential enzyme activity, and many factors such as gut environment, host diet, and intraspecific competition will play a role in bacterial community metabolic functions. Our results indicate that members of the Lachnospiraceae and Ruminococcaceae families that can potentially be important butyrate producers were decreased in NSCLC patients, which could be a factor contributing to immunotherapy responses. Although it is unlikely that the effect of the microbiome on ICI efficacy is the result of an individual family or species, it is instead the result of the whole microbiome modulating the host immune system. Nevertheless, certain groups of bacterial species could serve as predictive biomarkers of the treatment response in NSCLC patients [30].
A key strength of our study was that sample collection for the healthy control group and the treated NSCLC patients was undertaken within concurrent clinical studies in the same ethnic population and accounting for factors such as age, sex, BMI, and comorbidities such as cardiovascular disease, diabetes, and gastrointestinal conditions. Furthermore, the 16S-sequencing was performed in parallel for the control and the NSCLC group, thereby reducing errors in data collection.
Limitations of our study include the small sample size for the treated NSCLC patient group, which was only 18 individuals, and the fact that we did not obtain baseline fecal samples of NSCLC patients prior to treatment but instead utilized opportunistic sampling during the pembrolizumab treatment period or immediately following pembrolizumab treatment. Therefore, we could not determine whether the dysbiosis observed is due to the underlying cancer diagnosis [24], that is, the metastatic NSCLC, the previous platinum-based chemotherapy, and pembrolizumab immunotherapy that all patients received, or due to the progression on treatment. We can therefore only conclude broad differences in bacterial profiles between healthy individuals and NSCLC patients. Although patients were allowed to receive antibiotics during the treatment period, the average number of days between antibiotic treatment and sampling was 260 days. Only a single patient received antibiotics within a 60-day period prior to stool sampling; in this case, Meropenem 500 mg was administered 14 days prior to sampling and was excluded from the current analysis. Healthy controls were known not to have received antibiotics at least 45 days before fecal sampling. While antibiotics can disrupt gut bacteria composition, diversity, and metabolic activity [72, 73], the effect on the data presented here is limited due to the infrequent and intermittent use of antibiotics over an extended period of time (12 months) for the NSCLC group as part of their treatment. The gut microbiome is typically expected to recover to normal composition within 1.5 months of antibiotic treatment [74]. Finally, detailed dietary information is not available for the NSCLC patient group, which would have provided additional context for the observed composition of the gut microbiome, especially if their diet was modified due to their cancer diagnosis or treatment. In particular, we lack the data on the consumption of fiber and plant-based proteins, which provide the essential substrate for SCFA-producing bacteria versus the consumption of animal protein and saturated fats (Western diet), which promote dysbiosis and the growth of bacteria associated with metabolic disorders [75].
5 Conclusion
In this study, we show significant differences in the bacteriome of the pembrolizumab-treated NSCLC patients versus healthy controls. We found a decrease in the Firmicutes to Bacteroides ratio, an increase in the Bacteroidaceae relative abundance and a decrease in the family Lachnospiraceae and Ruminococcaceae relative abundance in NSCLC patients. This is consistent with previous studies which observe lower bacterial diversity, reduced beneficial bacteria such as SCFA producers, and an increase in potentially harmful bacteria in cancer patients receiving immunotherapy treatments. As a pilot study relying on opportunistic sampling of chemotherapy and immunotherapy treated metastatic NSCLC patients compared to healthy controls, we demonstrate that there is a large degree of dysbiosis in these patients, which from previous studies correlates with a compromised immune system and poor immunotherapy efficacy, as was in fact observed in this study (with only three patients long term survivors). These data are therefore hypothesis-generating for the need to address this degree of dysbiosis, by modulating the microbiome prior to or concurrent with treatment for metastatic NSCLC patients. We would however like to emphasize that our findings need to be confirmed in future prospective longitudinal studies with samples and dietary information at baseline from metastatic NSCLC patients prior to any treatment, and then during treatment to ascertain to what degree the dysbiosis seen is due to the metastatic NSCLC or the treatment or the progression on treatment, and to quantify the differences in the gut microbiome of NSCLC patients during immunotherapy and the role of specific bacteria in determining responses to immunotherapy.
Author Contributions
Haris Charalambous, Athos Antoniades: conceptualization, methodology, writing – original draft. Cameron Brown, Paris Vogazianos, Dimitris Vraxnos, Aristos Aristodimou: data analysis, interpretation, writing – original draft. Elisavet Papageorgiou, Ioannis Stylianou, Christos Shammas, Elpiniki Nikolaou, Jianxiang Chi, Paul Costeas: data curation, methodology, writing – review and editing. Kyriaki Katsaounou, Yiorgos Apidianakis: writing – review and editing.
Acknowledgments
The SWIPE clinical trial (NCT 02705820) was supported in part by a research grant from the Investigator-Initiated Studies Program of Merck Sharp & Dohme Corp. The opinions expressed in this paper are those of the authors and do not necessarily represent those of Merck Sharp & Dohme Corp.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.