Culture-enriched community profiling improves resolution of the vertebrate gut microbiota
Abstract
Vertebrates harbour gut microbial communities containing hundreds of bacterial species, most of which have never been cultivated or isolated in the laboratory. The lack of cultured representatives from vertebrate gut microbiotas limits the description and experimental interrogation of these communities. Here, we show that representatives from >50% of the bacterial genera detected by culture-independent sequencing in the gut microbiotas of fence lizards, house mice, chimpanzees, and humans were recovered in mixed cultures from frozen faecal samples plated on a panel of nine media under a single growth condition. In addition, culturing captured >100 rare bacterial genera overlooked by culture-independent sequencing, more than doubling the total number of bacterial sequence variants detected. Our approach recovered representatives from 23 previously uncultured candidate bacterial genera, 12 of which were not detected by culture-independent sequencing. Results identified strategies for both indiscriminate and selective culturing of the gut microbiota that were reproducible across vertebrate species. Isolation followed by whole-genome sequencing of 161 bacterial colonies from wild chimpanzees enabled the discovery of candidate novel species closely related to the opportunistic pathogens of humans Clostridium difficile and Hungatella hathewayi. This study establishes culturing methods that improve inventories and facilitate isolation of gut microbiota constituents from a wide diversity of vertebrate species.
1 INTRODUCTION
The gut microbial communities of vertebrates represent a significant reservoir of Earth's microbial diversity (Thompson et al., 2017) and can have profound effects on the phenotypes and fitness of their hosts (McFall-Ngai et al., 2013; Rosshart et al., 2017). However, most of the microbial diversity in the guts of vertebrates has never been cultured or isolated in the laboratory. The paucity of cultured biobanks limits the ability to phenotypically describe individual gut microbiota constituents, to sequence and assemble their entire genomes, and to experimentally decipher their relationships with one another and their hosts.
Recent work has developed effective approaches for culturing and generating isolate biobanks from the human gut microbiota (Browne et al., 2016; Lau et al., 2016; Poyet et al., 2019; Zou et al., 2019). For example, Lau et al. (2016) identified a panel of 33 media and two growth conditions capable of culturing >95% of the 16S rDNA 97% operational taxonomic units (OTUs) detected by culture-independent sequencing at >0.1% relative abundance in fresh human faecal samples. Similarly, Browne et al. (2016) showed that a single medium—yeast extract, casitone and fatty acid (YCFA)—cultured the majority of the bacterial species detected in fresh human faecal samples. These approaches represent exciting potential avenues for culturing the gut microbiota of diverse vertebrate species. However, the composition of the gut microbiota varies substantially among vertebrate species as a function of host phylogenetic history (Groussin et al., 2017; Ochman et al., 2010), diet (David et al., 2014; Muegge et al., 2011), physiology (Song et al., 2020), and geography (Moeller et al., 2017), and the extent to which the methods that have been developed to culture the human gut microbiota are applicable across vertebrate species remains poorly understood.
In addition, there is a lack of established methods for cultivating vertebrate gut microbiota from nonfresh samples. For many vertebrate species, the collection of fresh gut microbiota samples for anaerobic culturing is impractical. Samples are often collected in the field and stored in freezers, sometimes in a nucleic acid preservative such as ammonium sulphate solution (e.g., RNAlater). Recent efforts to culture the human gut microbiota have shown that a substantial fraction of humans’ faecal bacterial diversity can remain viable after freezing (Fouhy et al., 2015), but the resilience of gut microbiota to freezing has not been examined for most vertebrate species. Moreover, the cultivable fraction of the vertebrate gut microbiota from samples stored in nucleic acid preservatives has not been investigated.
Here, we cultured the gut microbiota of wild and captive chimpanzees, wild-derived inbred lines of house mice, wild-caught fence lizards, and humans from frozen faecal samples, including samples stored in RNAlater. Results showed that the majority of genera in the vertebrate gut microbiota are cultivable from frozen faecal samples under a single growth condition on a panel of nine media and that culturing revealed substantial gut microbiota diversity not readily detectable by culture-independent sequencing of 16S rRNA gene amplicons. In addition, we demonstrate how our approach can be used for both indiscriminate and selective isolation of bacterial lineages. Isolation of bacterial lineages from the gut microbiota of wild chimpanzees allowed whole genome sequencing of relatives of members of the human gut microbiota, including prominent opportunistic bacterial pathogens. Cumulatively, this study establishes effective methods for culturing the gut microbiotas of diverse vertebrate species from frozen faecal samples and demonstrates the utility of these methods for complete inventories of vertebrates’ gut microbiota diversity.
2 MATERIALS AND METHODS
2.1 Faecal collection
Faecal samples were collected from humans (Homo sapiens) (n = 2), captive chimpanzees (Pan troglodytes schweinfurthii) (n = 4), wild chimpanzees (n = 2) (Pan troglodytes schweinfurthii), wild-caught Western fence lizards (Sceloporus occidentalis) (n = 14), and wild-derived inbred mouse lines (Mus musculus domesticus) (n = 4). Human faecal samples were self-collected by two anonymous adult individuals from Ithaca, New York and frozen at –80°C within 1 h. Captive chimpanzee faecal samples were collected under direct observation from four individuals at Project Chimps, a sanctuary for retired research chimpanzees, and frozen at –80°C within 1 h. Faecal samples from wild chimpanzees were collected from two individuals at Gombe National Park and stored in RNAlater, then subsequently frozen at –80°C Faecal samples from wild-caught fence lizards were collected in the laboratory and frozen at −80°C as described by (Moeller et al., 2020). Faecal samples from two wild-derived inbred mouse lines (NY3 and NY4), which were derived from mouse populations in New York State and bred in the laboratory by sibling mating for >15 generations, following methods described by (Moeller et al., 2018), were collected under direct observation and frozen at –80°C within 1 h. All faecal collection was conducted with approval from Institutional Review Board of Cornell University and Institutional Animal Care and Use Committees of Cornell University and California Polytechnic State University.
2.2 Culturing microbiota from faecal samples and isolation of bacterial lineages
For each dry-frozen faecal sample, 0.1 g of material was diluted in 900 μl of brain heart infusion (BHI) broth (BD) with 0.05% l-cysteine hydrochloride hydrate (100 dilution). Then, 100 μl of 10−4 diluted samples were pipetted under anaerobic conditions onto a set of agar plates prereduced for ~12 h. Recipes for media are presented in Table S1. For wild chimpanzee faecal samples frozen in RNAlater, 100 μl of nondiluted samples were used. All plates were incubated at 37°C for five days in an anaerobic (5% hydrogen, 5% carbon dioxide and 90% nitrogen) chamber (Coy Lab Products Inc). After incubation, total cultures were collected from each plate by adding 500 μl BHI broth and scraping the surface of plates with a cell scraper (Corning Inc.). Plate scrapings (60 μl) were mixed with 320 μl of 50% glycerol and stored at –80°C to preserve cultured microbiota. The remaining volume of plate scrapings was transferred to a 1.5 ml Eppendorf tube and centrifuged for 1 min at 16,000 g to pellet down the bacteria for DNA extraction.
For wild chimpanzee faecal samples preserved in RNAlater, YCFA plates were cultured in duplicate, one of which was used for collecting total cultures and the other for picking single colonies. To isolate individual bacterial lineages, single colonies on YCFA plates from wild chimpanzees were picked using toothpicks, inoculated into YCFA broth and incubated for 2–3 days in an anaerobic chamber, shaking at 37°C. Isolated bacterial cells were then pelleted down at 3500 g for 10 min and stored at –20°C for DNA extraction.
2.3 16S rDNA amplicon sequencing
DNA was extracted from all faecal pellets and plate contents using a bead beating procedure and Qiagen PowerLyzer kit. For the human, wild chimpanzee, fence lizard, and house mouse samples, the V4–V5 region of 16S rDNA was amplified in duplicate with barcoded 515F 926R primer pair and the high fidelity Phusion polymerase as described previously (Comeau et al., 2017). For the captive chimpanzee samples, the V4 region of 16S rDNA was amplified in duplicate with barcoded 515F 806R primer pair as described previously (Walters et al., 2016) using the high fidelity Phusion polymerase. Previous validation wok has shown that, when combined with the universal 515F primer, 926R and 806R primers are expected to yield highly concordant results (Walters et al., 2016), and downstream analyses of culture media performance within each host species were not confounded with the reverse primer used for amplification. 16S rDNA libraries were pooled in equimolar amounts and sequenced on Illumina MiSeq lane using 300 + 300 bp paired-end V3 chemistry following protocols established by Comeau et al. (2017).
2.4 16S rDNA-amplicon sequence processing and analyses
Illumina reads from 16S rDNA amplicon sequencing were processed on the Qiita webserver (https://qiita.ucsd.edu/). Paired-end reads were filtered for quality and merged with Split libraries fastq using default settings and trimmed to a common length of 250 base pairs. ASVs were determined with Deblur (Amir et al., 2017) using default settings, and chimeras were removed from unrarefied data with removeBimeraDenovo in dada2 (Callahan et al., 2016).
All subsequent 16S rDNA analyses were conducted in qiime 2.0 (Bolyen et al., 2019). ASVs were inserted into the Greengenes reference phylogeny using fragment-insertion sepp. Taxonomy was assigned against the Greengenes (DeSantis et al., 2006) and silva (Quast et al., 2013) databases using classify-sklearn, and SILVA assignments were used for all downstream analyses unless otherwise specified. Each sample was rarefied to an even depth of 10,000 reads for all beta and alpha diversity analyses. Taxa barplots were constructed using barplot. Beta diversity between all pairs of samples were calculated with unweighted UniFrac (Lozupone & Knight, 2005), and alpha diversity of each sample was calculated with Faith's phylogenetic diversity (Faith, 1992). Significant differences between beta and alpha diversity among sample groups were assessed with PERMANOVA/PERMDISP (Anderson, 2001) and Kruskal-Wallis tests (Kruskal & Wallis, 1952) using group-significance. Adonis2 (Dixon & Palmer, 2003) with 999 permutations was employed to test for independent effects of host species and media on UniFrac dissimilarities among plate contents. Individual phylogenies of 16S rDNA sequences recovered from faecal samples and plate contents from each host group were constructed using align-to-tree-mafft-iqtree. The phylogenetic distributions of ASVs in faecal samples and plate contents were visualized with EMPress.
2.5 Extraction of genomic DNA from bacterial isolates
Genomic DNA was extracted from isolates in 96-well plates using the MagMAXTM CORE Nucleic Acid Purification Kit following manufacturers instructions. For mechanical disruption, 200 μl of sample was added to 450 μl of lysis solution in cluster tubes (Axygen, no. MTS-11-8-C-R) containing 0.5 mm diameter Zirconia/Silica beads (Biospec, no. 11079105z) and vortexed on a Mini-Beadbeater-96 (BioSpec, no. 1001). The samples were disrupted for 2 × 2.5 min, with a 5 min rest between disruptions. After mechanical disruption, the lysate was centrifuged for 5 min at 1048 g.
2.6 Library preparation and sequencing of isolate genomes
Whole-genome libraries from isolate DNA extractions were prepared using a modified Hackflex method (Gaio et al., 2019) with laboratory-made and adapted reagents from the Nextera DNA Flex Library Prep kit (Illumina). Briefly, BLT beads were diluted 1:40 with nuclease free water (Invitrogen), mixed with 10–50 ng of gDNA and 5 μl of 2× tagmentation buffer (20 mM Tris [pH 7.6] [Invitrogen], 20 mM MgCl [Sigma], and 50% [v/v] dimethylformamide [DMF] [Sigma]). Samples were incubated at 55°C for 15 min then held at 10°C, and 2.5 μl of 0.2% of sodium dodecyl sulphate (SDS; Sigma) was added to each sample. Samples were then incubated at 37°C for 15 min to stop tagmentation. Beads were then washed three times with 25 μl of washing solution (0.22 μm MF-millipore membrane filtered solution of 10% polyethylene glycol [PEG] 8000 [Sigma], 0.25 M NaCl [Sigma] in Tris-EDTA buffer [TE] [Invitrogen]). Libraries were prepared with PrimeSTAR GXL DNA Polymerase kit (Takara) following the manufacturer protocol. Each reaction contained 5 μl of 5× GXL buffer, 2 μl of 25 mM dNTPs, 1 μl of PrimeStar GXL polymerase, and 2 μl of nuclease free water. The PCR mix was added into washed beads and 5 μl of custom synthesized Oligos i5 and i7 added to a final concentration of 0.555 μM in each reaction. Amplification was performed as follows: 3 min at 68°C, 3 min at 98°C, 12 cycles of (45 s at 98°C–30 s at 62°C–2 min at 68°C), 1 min at 68°C and hold at 10°C. Libraries were incubated at room temperature with paramagnetic beads for 5 min, washed twice with 200 μl fresh 80% ethanol, and then resuspended in 30 μl of ultrapure water (Invitrogen) and transferred into a new tube. The concentrations and fragment sizes of eluted libraries were measured using Qubit high sensitivity (HS) dsDNA kit (Thermo Fisher Scientific) and the HS Bioanalyzer chip (Agilent), respectively. Final isolate genome libraries were pooled and sequenced on a single P2 Illumina NextSeq 2000 flow cell at the Cornell Institute of Biotechnology.
2.7 Assembly of isolate genomes
Isolate sequences were processed, assembled, and annotated with the Bactopia genome analysis pipeline (Petit & Read, 2020) using default parameters. Bactopia employs the Nextflow workflow management system (Di Tommaso et al., 2017) to efficiently process raw sequence data. The genome assemblies presented in this manuscript were assembled using the skesa assembler (Souvorov et al., 2018) from sequences trimmed with BBduk (sourceforge.net/projects/bbmap). Assembly quality was assessed with quast (Gurevich et al., 2013), and completeness and contamination assessed using checkm (Parks et al., 2015) using default parameters. Isolate assemblies displaying >10% contamination were excluded from downstream analyses.
2.8 Classification and phylogenomics of chimpanzee-derived bacterial isolates
Bacterial lineages isolated from chimpanzees were classified to taxonomic groups with the Genome Taxonomy Database Toolkit (GTDB-Tk) using default parameters. To evaluate the relationships of bacteria isolated from chimpanzees and constituents of the human gut microbiota, we identified for each chimpanzee-derived genome the most similar genomes represented in the Human Microbiome Project (HMP) isolate collection. MinHash sketches of all chimpanzee-derived genomes and all isolate genomes generated by the HMP were compared using Mash (Ondov et al., 2016) to identify for each chimpanzee-derived genome the top five most similar genomes in the HMP collection.
Anvi'o was employed to generate an alignment of concatenated single copy proteins present in the Bacteria_71 collection from all chimpanzee-derived bacterial genomes and their top-5 closest MASH hits from the HMP collection. A maximum likelihood phylogeny with 100 bootstrap replicates was inferred based on the concatenated alignment using RAxML under the PROTGAMMWAG model of substitution.
3 RESULTS
3.1 Taxonomic composition of microbiota in faecal samples and cultures from vertebrate species
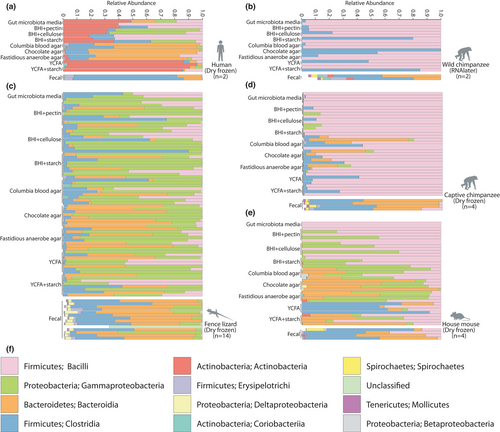
The workflow for culture-enriched molecular profiling (Lau et al., 2016) is presented in Figure 1. Faecal samples from humans (Homo sapiens) (n = 2), captive chimpanzees (Pan troglodytes schweinfurthii) (n = 4), wild chimpanzees (P. t. schweinfurthii) (n = 2), wild-caught Western fence lizards (Sceloporus occidentalis) (n = 14), and wild-derived inbred mouse lines (Mus musculus domesticus) (n = 4) were cultured on a panel of 10 media, including nine media that were used for all host species (Table S1). The V4–V5 region of the 16S rDNA gene was amplified from all faecal samples (n = 26) and contents of culture plates (n = 190). Amplicons were sequenced on an Illumina MiSeq, generating 6,960,630 high-quality sequences representing 18,415 amplicon sequence variants (ASVs). The number of reads per sample ranged from 3609 to 92,631, with a median of 32,225 reads per sample. The rarefaction depth for analyses of alpha and beta diversity of 10,000 reads was obtained for all but three samples. A list of samples and their corresponding metadata are presented in Table S2.

Taxonomy assignments of reads against Greengenes version 13.8 and Silva version 132 databases indicated that faecal samples were dominated by Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. These phyla were also predominant on culture plates. Stacked bar charts of the relative abundances of bacterial classes based on Greengenes assignments are presented in Figure 2. Greengenes and Silva taxonomic assignments of ASVs are presented in Table S3.
3.2 Culturing captures vertebrate gut microbiota diversity not detected by culture-independent sequencing
To determine what fraction of microbial taxa in each host species’ faecal samples were cultivable, we identified taxa detected only in faecal samples by culture-independent sequencing, only in plate contents, or in both faecal samples and plate contents. Because the total number of ASVs observed may be inflated by sequencing errors, we conducted analyses of both ASVs and higher taxonomic ranks ranging from 97% OTU to phylum (Table 1). In total, we detected 340 bacterial genera in faecal samples, 299 bacterial genera in cultured plate contents, and 184 bacterial genera in both faecal samples and plate contents (Table S4). Of the 299 bacterial genera detected in culture plate contents, 24 were labelled as “uncultured” candidate genera in taxonomic databases. Similarly, of the 115 bacterial genera detected only in culture, 12 were labelled as “uncultured” candidate genera in taxonomic databases. The phylogenetic distributions of ASVs from each host species detected in faecal samples, detected in plate contents, or detected in both faecal samples and plate contents are presented in Figures 3 and S1–S5.
| Taxonomic rank | Fence lizard | House mouse | Wild chimpanzee | Captive chimpanzee | Human |
|---|---|---|---|---|---|
| ASV | |||||
| Culture only | 2739 | 1353 | 468 | 1739 | 1306 |
| Faecal only | 1902 | 996 | 893 | 4223 | 663 |
| Both | 236 | 98 | 43 | 134 | 109 |
| OTU | |||||
| Culture only | 678 | 466 | 154 | 790 | 281 |
| Faecal only | 595 | 713 | 495 | 2755 | 442 |
| Both | 149 | 61 | 33 | 157 | 82 |
| Genus | |||||
| Culture only | 121 | 63 | 42 | 43 | 86 |
| Faecal only | 96 | 44 | 116 | 173 | 53 |
| Both | 63 | 56 | 38 | 89 | 72 |
| Family | |||||
| Culture only | 79 | 42 | 13 | 18 | 43 |
| Faecal only | 39 | 11 | 45 | 65 | 4 |
| Both | 35 | 35 | 19 | 37 | 34 |
| Order | |||||
| Culture only | 44 | 36 | 19 | 9 | 48 |
| Faecal only | 23 | 4 | 0 | 0 | 1 |
| Both | 26 | 19 | 8 | 42 | 14 |
| Class | |||||
| Culture only | 19 | 11 | 0 | 4 | 10 |
| Faecal only | 8 | 4 | 13 | 17 | 1 |
| Both | 20 | 15 | 14 | 13 | 13 |
| Phylum | |||||
| Culture only | 9 | 7 | 0 | 3 | 7 |
| Faecal only | 7 | 3 | 6 | 11 | 1 |
| Both | 11 | 8 | 9 | 8 | 6 |

3.3 Chocolate agar outperforms other media in culturing faecal microbiota from each host species
To evaluate the performance of each media for culturing the microbiota of each host species, we calculated UniFrac dissimilarities, which measure the overlap between the phylogenetic trees of two microbial communities, for each pair of samples in our data set. The cultured microbiota from each host species was distinguishable from the cultured microbiota of every other host species. For every host species, microbiota in plate contents cultured from the same host species were more similar on average than microbiota in plate contents cultured from different host species (Figures S6 and S7).
A single media, chocolate agar (CA), best cultured the bacterial diversity detected in faecal samples from each host species, including both dry frozen faecal samples and faecal samples frozen in RNA later. The mean UniFrac dissimilarity between CA plate contents and host faecal samples was less than that of any other media for each host species (Figure 4) (nonparametric permutation test p = .001). CBA also consistently performed well, whereas the worst performing media varied across host species (Figure 4). YCFA and gut microbiota media, which have been widely used in efforts to culture the human gut microbiota (Browne et al., 2016; Forster et al., 2019; Goodman et al., 2011; Ito et al., 2019; Yousi et al., 2019), both performed worse than CA and CBA for each host species.
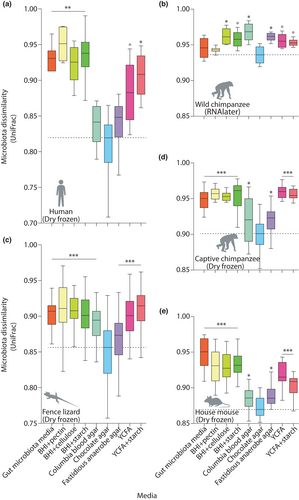
CA and CBA also cultured the overall highest levels of alpha diversity of any media for each host species. The alpha diversities, calculated as Faith's phylogenetic diversity, for each combination of media and host species are presented in Figure S8. Alpha diversity rarefaction curves for plates from each host species are presented in Figure S9.
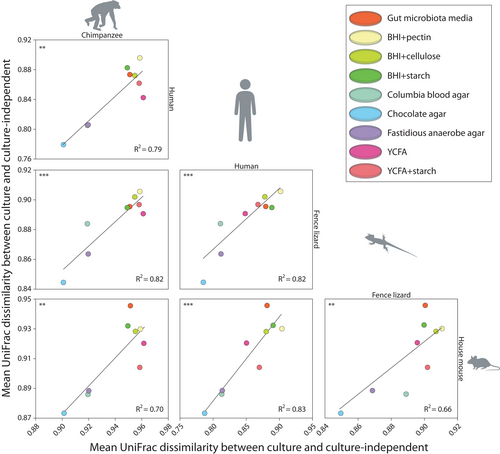
3.4 Reproducible performance rankings of media for culturing gut microbiota across host species
To evaluate whether the culturing performances of media were consistent across host species, we tested whether UniFrac dissimilarities between plate contents from each media and faecal samples were associated between each pair of host species. These analyses indicated significant consistency across host species in the degree to which specific media cultured the gut microbiota diversity detected in faecal samples. The mean UniFrac dissimilarities between media and faecal samples were positively associated for each pair of host species (Figure 5) (p-value for nonzero slop <.01 in each comparison). The p-values and other statistics for these regression analyses are presented in Table S5. In addition, results of core microbiome analyses conducted in QIIME2 identified sets of bacterial genera consistently recovered by specific media across host species (Supporting Information, Table S6, Table S7).
3.5 Media select for distinct sets of gut bacterial taxa from each host species
Adonis2 analyses indicated that the composition of the microbiota cultured by each plate depended on the type of media used as well as the host species from which the faecal sample was cultured. The variables media and host species independently explained a significant portion of UniFrac beta-diversity among plate contents. The variable media explained 12.7% of the variation independently of host species (nonparametric permutation test p = .001), whereas the variable host species explained 12.9% of the variation in microbiota beta diversity among plate contents independently of media (nonparametric permutation test p-value = .001). In addition, PERMANOVA/PERMDISP indicated significant differences in beta-diversity centroids between host species (Supporting Information). To enable informed selection of media for the cultivation of specific bacterial taxa by future studies, the ASVs detected by different combinations of media and host species are presented in Table S8. In addition, a heatmap of bacterial classes detected in each faecal sample and culture plate is presented in Figure S10.
3.6 Culturing microbiota from RNA-later preserved faecal samples and isolate whole-genome sequencing
Faecal samples from wild chimpanzees preserved in RNAlater, an aqueous ammonium sulfate storage reagent, were the most recalcitrant to culture. These faecal samples displayed the highest UniFrac dissimilarities from their cultures of any faecal samples cultured (Figure 3). In addition, cultures from these faecal samples displayed the lowest alpha diversity of any cultures (Figure S8).
However, certain bacterial lineages were consistently detected in culture from faecal samples preserved in RNAlater. The cultivable fraction of RNAlater preserved faecal samples spanned several phyla and included predominant members of microbiota within the Clostridialles, Bacillales, and Enterobacterales. Additional experiments using two media (YCFA and YCFA+starch) and RNAlater preserved faeces from wild living bonobos (Pan paniscus) and western chimpanzees (Pan troglodytes troglodytes) (Table S9) also indicated the cultivability of these lineages from samples stored up to 16 years (Supporting Information, Figure S11). Similarly, analyses using the QIIME implementation of SourceTracker (Knights et al., 2011) indicated that these results could not be attributed to DNA contamination from faecal samples (Figure S12).
To confirm the viability of these cultured lineages, we isolated 161 colonies cultured from wild chimpanzee faecal samples preserved in RNAlater and sequenced their genomes. SPAdes (Bankevich et al., 2012) assembly statistics for each genome calculated in QUAST (Gurevich et al., 2013) are presented in Table S10. Of these genomes, 142 contained sequences from over 50% of the genes in the Anvi'o Bacteria_71 universal single-copy protein-coding gene collection. Similarly, CheckM results indicated that 142 genomes were over 90% complete, with 110 being over 90% complete and less than 10% contaminated. The 16S rDNA sequences from each genome identically matched 16S rDNA sequences detected in culture, indicating the viability of these lineages after several years of frozen storage in RNAlater. The 16S rDNA sequences from each genome are presented in File S1. Taxonomy analyses using GTDB-Tk indicated that these 110 high-quality isolate genomes included 13 genomes belonging to putatively novel species (Supporting Information).
3.7 Isolate genomes from wild chimpanzees represent close relatives of the human gut microbiota
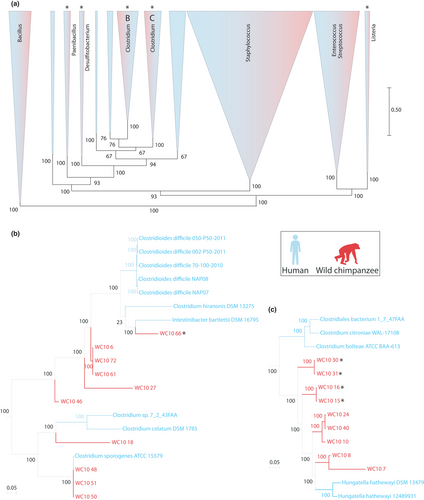
Previous work has shown that the gut microbiota of wild chimpanzees contains close relatives to members of the human gut microbiota, some of which have codiversified with host species (Moeller et al., 2016). To determine the relationships between the 142 chimpanzee-derived bacterial genomes that we sequenced and human-derived gut bacterial genomes, we identified using Mash (Ondov et al., 2016) the top five closest sequence matches from the Human Microbiome Project isolate collection for each chimpanzee-derived bacterial genome. Next, we used RAxML (Stamatakis, 2006) to construct a maximum-likelihood phylogeny of all chimpanzee-derived and human-derived bacterial genomes based on an alignment of Anvio's Bacteria_71 universal single-copy protein-coding genes. These analyses revealed that chimpanzee-derived bacteria represent distinct lineages that are closely related to members of the human gut microbiota, including the human pathogens Clostridium difficile, Hungatella hathewayi, and Listeria spp. as well as commensal Enterobacter and Desulfitobacterium spp. (Figures 6 and S13). All isolates are available upon request.
4 DISCUSSION
We found that representatives from over half of the bacterial genera in the gut microbiotas of chimpanzees, house mice, fence lizards, and humans were cultivable from frozen faecal samples on a panel of nine media under a single growth condition (Table 1). In addition, culturing allowed the detection of low-abundance lineages not observed by culture-independent sequencing. Cultured lineages included 297 bacterial genera. These genera included 23 previously uncultured candidate bacterial genera, 12 of which were not detected by culture-independent sequencing (Table S4). Because the same sequencing approach was applied to both faecal samples and agar plate contents, the observation that 12 genera were detected in culture but not in faecal samples cannot be explained by sequencing bias. Instead, this result suggest that culturing amplifies members of the rare biosphere within the vertebrate gut microbiota, revealing lineages missed by culture-independent approaches. Overall, culturing more than doubled the number of 16S rDNA amplicon sequence variants (ASVs) detected from the gut microbiotas of vertebrate host species. These results indicate that our culturing approach (i) recovers a substantial fraction of the gut microbiota of diverse vertebrate species, including previously uncultured and unclassified taxa, and (ii) provides more complete inventories of bacterial diversity within vertebrate faecal samples relative to culture-independent sequencing alone.
We observed striking consistency among vertebrate species in the degree to which specific media successfully cultured the microbiota diversity present in faecal samples (Figures 3 and 4). For example, CA and CBA cultivated microbiota that most closely matched faecal microbiota for each vertebrate species. These media also cultured the highest levels of alpha diversity of any media for each vertebrate species (Figure S8), although estimates of alpha diversity were lower in culture than in faecal samples. CA and CBA are compositionally similar, both containing mammalian blood products. Blood agars facilitate the growth of fastidious microorganisms and have been shown to effectively culture diverse members of the human gut microbiota (Lagier et al., 2015). Here, we did not add blood to Fastidious Anaerobe Agar, which may have contributed to the observation that this media performed worse than CA and CBA. In contrast to blood agars, other media that we examined cultivated more specific subsets of the microbiota (Figure S10; Table S6). For example, brain heart infusion with pectin (BHI+pectin) performed the worst of all media examined in terms of recapitulating bacterial diversity present in faecal samples. Relative to all other media, BHI +pectin grew bacterial communities that were the least compositionally similar to those present faecal samples for each vertebrate species. However, BHI +pectin consistently recovered many of the same gut bacterial taxa across all vertebrate species (Figure S10; Table S6). Compared to blood agars, these relatively selective media will be more applicable for future culturing efforts targeting subsets of lineages within the vertebrate gut microbiota.
Similarly, bacterial communities grown on YCFA displayed greater dissimilarities to faecal samples than those grown on blood agars, despite previous results indicating that YCFA is capable of supporting the growth of most of the bacterial species in human faecal samples detected by metagenomic sequencing (e.g., Browne et al., 2016). This discrepancy could reflect an interaction between sample freezing and media performance. For example, freezing faecal samples may enrich for certain lineages present in only low abundances in fresh samples. If those lineages enriched by freezing competitively exclude other lineages, this effect could reduce the performance of the media for frozen samples relative to fresh samples. Previous work has shown that freezing alters the cultivability of the human gut microbiota (Fouhy et al., 2015; Lau et al., 2016). Expanding upon our results, future work will benefit from direct comparisons of media performance between frozen and fresh samples across vertebrate species for which fresh samples are available.
We found that representatives from the majority of bacterial genera in the gut microbiota can be cultured from frozen faecal samples. However, our study provides only a lower bound estimate of the true cultivability of the gut microbiota from frozen samples. It is likely that additional atmospheric conditions (microaerophilic, aerobic), additional media, and longer cultivation periods would allow the cultivation of additional gut microbiota diversity than that detected here. For example, although Lau et al. (2016) showed that >99% of the bacterial 16S rDNA OTUs detected in human faecal samples could be recovered on culture plates using a 5-day growth period, Lagier et al. (2016) found that additional gut microbiota diversity continues to appear on plates over culturing periods of multiple weeks. These results indicate that comprehensive culturing of bacterial diversity present in frozen faecal samples would require additional growth conditions and durations than those examined here. Nevertheless, our results establish a reproducible and relatively simple approach for cultivating gut microbiota from a diversity of vertebrate species from frozen faecal samples requiring only a single growth condition (anaerobic), <10 media, and a <1 week cultivation period.
Another limitation of the current study that may be addressed by future work is the identification of cultivable and uncultivable strain-level variation in the vertebrate gut microbiota. We found that representatives from the majority of genera could be detected in mixed culture, but the proportion of cultivable lineages was reduced at lower taxonomic levels (e.g., ASV level). The application of metagenomic shotgun sequencing, rather than the 16S rDNA amplicon sequencing performed here, is expected to provide increased resolution of the culturable and unculturable subspecies lineages from frozen faecal samples.
In addition to demonstrating the cultivability of the gut microbiota from dry frozen vertebrate faecal samples, we found that certain gut bacterial lineages were cultivable from frozen faecal samples preserved in ammonium sulphate solution (i.e., RNAlater) (Figures 2, 6, and S11). These samples from wild chimpanzees were frozen in RNAlater for >4 years (>16 years for some samples, Table S9), indicating that certain members of the gut microbiota remain viable in this preservative despite long-term storage. Compared to plates cultured from dry-frozen faecal samples, the plates cultured from RNAlater-preserved faecal samples contained less diverse microbial communities (Figure S8). In addition, the plate contents from these samples were the least similar of any cultured plate contents to the microbiota composition inferred from culture independent sequencing of their corresponding faecal samples (Figure 4). These results indicate that RNAlater preserves a relatively small fraction of the gut microbiota, suggesting that this preservative can be used as a selective filter for targeted culturing of specific bacterial clades. This observation reflects previous work indicating that ethanol and acid treatments can be used as a selective filter for certain resilient bacterial taxa, such as spore formers (e.g., Browne et al., 2016). However, additional experiments in which individual faecal samples are aliquoted and either dry frozen or frozen in RNAlater will be required to identify specific bacterial lineages that are affected by the different storage conditions.
In addition to improving detection of gut microbiota diversity, culturing also enabled the isolation of individual gut bacterial lineages. Recent studies have generated gut bacterial biobanks and whole-genome references databases from humans (Browne et al., 2016; Forster et al., 2019; Lagier et al., 2016; Poyet et al., 2019; Zou et al., 2019), laboratory mice (Liu et al., 2020), and agricultural ruminants (Seshadri et al., 2018). The approaches we describe here will allow the creation of similar biobanks of the gut microbiota from a wide diversity of vertebrate hosts, including wild living hosts for which only frozen faecal samples are available. It is likely that many of the bacterial lineages that we detected in mixed cultures on agar plates cannot be isolated (e.g., due to their reliance on other members of the microbiota), but whole-genome sequencing indicated that a substantial proportion of colonies that we picked from plates represented individual isolates. For example, whole genome sequencing of 161 bacterial colonies from the gut microbiota of wild chimpanzees revealed a diverse collection of isolates, including lineages closely related to known pathogens in the human gut microbiota (Figure 6). The isolates from a wild chimpanzee included lineages closely related to the opportunistic pathogens of humans Hungatella hathewayi and Clostridium difficile (Figures 6 and S13), the latter of which is the leading bacterial cause of hospital-acquired infections (Magill et al., 2014). Several of these lineages represent putatively novel bacterial species (Figures 6 and S13) belonging to taxa previously reported to reside within the wild chimpanzee gut microbiota (Moeller et al., 2012, 2016). The development of isolate biobanks from chimpanzees and other apes—humans closest living relatives—will provide an invaluable resource for investigating the evolutionary history of the human gut microbiota, including the evolution of pathogenicity within otherwise commensal bacterial clades. Methods for culturing the gut microbiota of wild vertebrates may have conservation implications. The gut microbiota of vertebrates can be disrupted by anthropogenic activities both in the wild (Amato et al., 2016; Gomez et al., 2015) and in captivity (Clayton et al., 2016; McKenzie et al., 2017) with potential consequences for health. For example, compositional differences in the gut microbiota between captive and wild black rhinoceros may be contributing to digestive issues in captive populations (Gibson et al., 2019). These observations have led to interest in considering the gut microbiota in conservation strategies for threatened or endangered vertebrates (Gibson et al., 2019; Trevelline et al., 2019). Our observations that a substantial fraction of the vertebrate gut microbiota is cultivable from frozen faecal samples indicate that gut microbiota biobanks can be generated from wild-living vertebrate species even if fresh faecal samples for anaerobic culturing are not available. Culturing and biobanking the gut microbiota of vertebrate species, many of which harbor unique gut microbial lineages found in no other habitat (Thompson et al., 2017), could facilitate efforts to restore the gut microbiota of vertebrates affected by human-mediated dysbiosis.
ACKNOWLEDGEMENTS
We thank Dan Sprockett for assistance with data analysis. For permission and support to conduct research on wild chimpanzee, we are grateful to the Jane Goodall Institute, the Gombe Stream Research Centre, Tanzania National Parks, Tanzania Commission for Science and Technology, and the Tanzania Wildlife Research Institute. This work was supported by grants from the National Institutes of Health (R35 GM138284, R01 AI120810, R01 AI050529, and R01 AI150590), Cornell University College of Agriculture and Life Sciences, the US Fish and Wildlife Great Ape Conservation Fund, the Arcus Foundation, and the Leo S. Guthman Foundation.
AUTHOR CONTRIBUTION
Andrew H. Moeller conceived and designed the research plan. Jon G. Sanders and Weiwei Yan performed the laboratory work. Anthony Denice, Margaret Cornwall, Kathleen N. Ivey, Emily N. Taylor, Alex R. Gunderson, Michael J. Sheehan, Deus Mjungu, Elizabeth V. Lonsdorf, Anne E. Pusey, Beatrice H. Hahn, and Andrew H. Moeller provided samples. Samantha L. Goldman, Jon G. Sanders, and Andrew H. Moeller analysed the data. Samantha L. Goldman, Jon G. Sanders, Weiwei Yan, and Andrew H. Moeller wrote the manuscript.
CONFLICT OF INTERESTS
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
All 16S rDNA and genomic sequences have been deposited in the European Nucleotide Archive under accession PRJEB44552. Data are also available at qiita.ucsd.edu under study ID 13042. All bacterial isolates are available upon request (Goldman et al., 2021).




