DomeTree: a canonical toolkit for mitochondrial DNA analyses in domesticated animals
Abstract
Mitochondrial DNA (mtDNA) is widely used in various genetic studies of domesticated animals. Many applications require comprehensive knowledge about the phylogeny of mtDNA variants. Herein, we provide the most up-to-date mtDNA phylogeny (i.e. haplogroup tree or matrilineal genealogy) and a standardized hierarchical haplogroup nomenclature system for domesticated cattle, dogs, goats, horses, pigs, sheep, yaks and chickens. These high-resolution mtDNA haplogroup trees based on 1240 complete or near-complete mtDNA genome sequences are available in open resource DomeTree (http://www.dometree.org). In addition, we offer the software MitoToolPy (http://www.mitotool.org/mp.html) to facilitate the mtDNA data analyses. We will continuously and regularly update DomeTree and MitoToolPy.
Introduction
Mitochondrial DNA (mtDNA) is a useful molecular marker for constructing matrilineal genealogies and tracing evolutionary history of domesticated animals from a matrilineal perspective (Bruford et al. 2003; Wang et al. 2014). It is also valuable for studies of ancient DNA (Ho & Gilbert 2010) as well as non-human forensic research (Imes et al. 2012). Since 2006, the number of complete mtDNA nucleotide sequences (i.e. mitochondrial genomes) has grown quickly due to the improved sequencing technologies and the high-quality commercial kits (Wang et al. 2014).
A high-resolution mtDNA phylogeny provides an essential framework for various studies and yet different approaches, such as Bayesian inference and maximum-likelihood trees, are used to infer historical relationships. The phylogeny as well as the nomenclature of mtDNA lineages fluctuates along with the accumulation of more data even for the same animal such as pig (Wu et al. 2007a,b). This phenomenon complicates the comparison of results from different studies especially when this involves different and updated nomenclatorial systems. Predominant in molecular anthropology, a phylogenetic network-like (Bandelt et al. 1999, 2000) mtDNA haplogroup tree successfully captures the known mtDNA variation (Torroni et al. 2006). In this system, mtDNA sequences are aligned against a reference sequence and the variants are then scored (Bandelt et al. 2014). A haplogroup is a cohort mtDNA lineages derived by descent from the same ancestral mtDNA molecule as revealed by the sharing of a characteristic mutational motif (i.e. strings of variants) (Torroni et al. 2006). Thus, a haplogroup represents a monophyletic clade (Wu et al. 2007b) and could be read from the mtDNA haplogroup tree (Torroni et al. 2006). Nowadays, the global human mtDNA haplogroup tree is maintained as PhyloTree (http://www.phylotree.org) (van Oven & Kayser 2009). It facilitates the comparison of different mtDNA studies. Some bioinformatic tools are based on this de facto standard (e.g. Fan & Yao 2011, 2013; Vianello et al. 2013).
The haplogroup tree-based strategy has been employed for pigs (Wu et al. 2007b), cattle (Achilli et al. 2008, 2009; Bonfiglio et al. 2010, 2012), horses (Achilli et al. 2012), chickens (Miao et al. 2013), sheep (Lancioni et al. 2013) and dogs, goats and yaks (Shi et al. 2014). Nevertheless, the established phylogeny including the haplogroup nomenclature (e.g. Wu et al. 2007b) is neglected in subsequent researches. Some recently released mtDNA sequences have not been analysed under such context. Different haplogroup/clade/group nomenclature has been proposed (e.g. Yu et al. 2013; Cannon et al. 2015). The generated confusion and chaos complicates the comparison of different studies. Therefore, we present an integrated bioinformatic approach to provide (i) the standardized mtDNA phylogeny with haplogroup nomenclature system; and (ii) the related software for data analysis, to address issues in the mtDNA studies of domesticated animals.
Materials and methods
Data sets
We only considered animals (including domesticates and their wild ancestors) with at least 20 complete or near-complete mtDNA genomes deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank/; Accessed on: June 1st, 2014). The data set includes a total of 1342 mtDNA sequences after scrutiny of data quality (Shi et al. 2014). Eight sequences (Table S1, Supporting information) fell outside the macro-clades (i.e. macro-haplogroups) containing other samples of dogs (Thalmann et al. 2013), pigs (Wu et al. 2007b) and yaks (Wang et al. 2010), respectively. More than 400 variants (data are not shown) were scored out for the sequence EU442884 of Mongolian wolf (Canis lupus chanco). Thus, a total of nine sequences (Table S1) with remote relationships to others as suggested before (Shi et al. 2014) were removed in subsequent analyses. For 194 sequences with potential errors (Shi et al. 2014), we (i) rescued 101 sequences (Table S2, Supporting information) by disregarding error-prone sites in next-generation sequencing (Table S3, Supporting information); and (ii) yet excluded the remaining 89 flawed sequences (Table S4, Supporting information). Ultimately, we used 1240 complete or near-complete mtDNA genome sequences.
mtDNA haplogroup trees
Sequences were aligned with SeqMan Pro of DNASTAR lasergen 7.1.0 (DNAStar Inc., Madison, WI) against reference sequences (Table 1). The mtDNA sequence alignments were deposited in Dryad (doi:10.5061/dryad.cc5kn). Sequences with different start positions were reassembled in terms of the corresponding reference sequences and then were realigned. Mutations were scored relative to the related reference sequences (Table 1). Difficult-to-align regions and error-prone sites in next-generation sequencing were excluded from further consideration. We followed the rules proposed by Bandelt and Parson (Bandelt & Parson 2008) to score length variants. We employed the matching or near-matching strategy (Yao et al. 2004) to screen haplogroup-specific mutations defined in available mtDNA haplogroup trees for each kind of domesticated animals (Wu et al. 2007b; Achilli et al. 2008, 2009, 2012; Bonfiglio et al. 2010, 2012; Lancioni et al. 2013; Miao et al. 2013; Shi et al. 2014) in the scored variants of every sequence. And then we could allocate each of sequences into specific haplogroup. As a result, the phylogenetic status for each of sequences could be determined so that the trees could be updated by incorporating all of sequences. The trees were checked by network 4.611 (http://www.fluxus-engineering.com/sharenet.htm) (Bandelt et al. 1999). The mtDNA haplogroup trees for domestic animals were transcribed into the extensible stylesheet language format (xls) as adopted by rCRS-oriented version of PhyloTree (http://phylotree.org/rCRS-oriented_version.htm) (van Oven & Kayser 2009) and were then deposited in dometree (http://www.demotree.org). The hierarchical haplogroup nomenclature systems were updated from previous studies (Table 1) while referring to the rules used in the phylogeny of human mtDNA (van Oven & Kayser 2009) in order to avoid chaos and confusion.
| Animals | Related species | No. of sequences | Reference sequence | Haplogroup nomenclature |
|---|---|---|---|---|
| Cattle and Aurochs | Bos taurus | 258 | V00654 | I, P, Q, R, and T |
| B. indicus | 9 | |||
| B. primigenius | 2 | |||
| Yak and wild yak | B. grunniens | 70 | GQ464259 | A–D |
| Dog and grey wolf | Canis lupus | 424 | EU789787 | A–F |
| Horse and Przewalski's horse | Equus caballus | 230 | JN398377 | A–R |
| E. przewalskii | 6 | |||
| Chicken and Red Junglefowl | Gallus gallus | 65 | AP003321 | A–I and X–Z |
| Pig and wild boar | Sus scrofa | 100 | EF545567 | A, D, and E |
| Sheep | Ovis aries | 45 | AF010406 | A–E |
| Goat | Capra hircus | 31 | GU068049 | A–C |
Application software
We developed mitotoolpy (http://www.mitotool.org/mp.html) written in Python language to handle data analyses. It was based on biopython (http://www.biopython.org/) (Cock et al. 2009) and licensed under gpl v3. The pairwise sequence alignment was based on clulstalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Larkin et al. 2007), as the results were similar to commercial software seqman pro of which codes were not available. The scoring system for mtDNA haplogroup determination was employed from our previously presented mitotool (Fan & Yao 2013). And the software fixed the issues of haplogrouping (Bandelt et al. 2012). mitotoolpy has been and will be updated synchronously with dometree.
Results
The updated mtDNA haplogroup trees
By analysing 1240 complete or near-complete published mtDNA genomes (Table 1), we updated the mtDNA haplogroup trees of cattle, dogs, goats, horses, pigs, sheep, yaks and chickens and deposited them in DomeTree (Fig. 1a). Monophyly of each major clades/haplogroups described in previous studies (Table 1) was confirmed. The diagnostic motif (in both control- and/or coding-region) for each (sub)haplogroup was characterized and mapped on each branch. All trees were made available for viewing with common web browsers such as Internet Explorer and Mozilla Firefox. It was convenient to use the ‘Find’ command of the browsers to search for specific haplogroups or mutations.
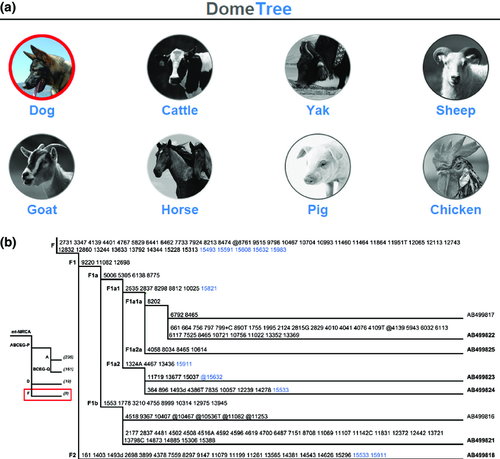
The incorporation of recently released sequences into our analyses defined some new (sub)haplogroups. In one case, sequences AB499818, AB499821-AB499825 of the extinct Japanese Honshu wolf (Canis lupus hodophilax) clustered with Kishu (AB499816) and Husky (AB499817) in haplogroup F (Fig. 1b; http://www.dometree.org/trees/dog.htm), which previously was only defined by control region information (Pang et al. 2009). In another case, sequence EF375877 of the Lanyu pig from Lanyu Islet (Wu et al. 2007a) rooted at the base of the branch for haplogroup A1, and, accordingly, was used to define new subhaplogroup A1c (http://www.dometree.org/trees/pig.htm).
Phylogenetic analyses of mtDNA data
In terms of mtDNA haplogroup trees, we developed MitoToolPy for data analyses, which included the following functions: (i) score variants relative to a reference sequence; (ii) classify haplogroups (haplogrouping); and (iii) check for potential errors due to missing diagnostic mutations as well as excessive private mutations. We applied mitotoolpy to 34 novel sequences released by GenBank (Table S5, Supporting information) and further confirmed the results manually. This trial found mitotoolpy to be practical and efficient. Moreover, mitotoolpy handled mtDNA fragments (e.g. control region) and served as a convenient ‘barcoding tool’ for some strains or breeds.
Discussion
The updated mtDNA phylogeny including its haplogroup nomenclature can serve as a starting point for future mtDNA analyses for cattle, dogs, horses, pigs, yaks and chickens. This is necessary for avoiding conflicts and confusion when comparing different mtDNA studies, especially now when next-generation sequencing accelerates the accumulation of mitochondrial genome data (Lippold et al. 2011; Horsburgh et al. 2013). The highly resolved, updated phylogenies often highlight the multiple matrilineal backgrounds of domesticated animals. For example, the mtDNA phylogeny of dogs and grey wolves indicates that haplogroup F (Fig. 1b) is a genetic relic of extinct Japanese Honshu wolves or their ancestors in modern Kishus and Huskies. Similarly, the haplogroup A1c in the Lanyu pig probably traces back to local wild boar. Similar scenarios exist for haplogroups P, Q and R in cattle (Achilli et al. 2008, 2009; Bonfiglio et al. 2010). Sporadic introgression from wild ancestors or local domestication events best explained these patterns (Wang et al. 2014). Greater effort, such as studies of ancient DNA, can shed light on such observations (Larson & Burger 2013). In addition, our results support that there are no signals of recombination neither in the phylogeny explored nor on maternal inheritance (Bandelt et al. 2005).
We will simultaneously update both dometree and mitotoolpy at least every year. Updates will reconcile issues of phylogeny and haplogroup nomenclature due to novel sequence data. Further, we plan to incorporate additional domesticated animals into this system as their data become accessible. We hope that dometree and mitotoolpy will facilitate and foster future work on the mtDNA of domesticated animals.
Acknowledgements
This work was supported by the 973 program (2013CB835200 and 2013CB835204). M-S.P. thanks the support from the Youth Innovation Promotion Association, Chinese Academy of Sciences. R.W.M. thanks the support from the Visiting Professorship for Senior International Scientists from the Chinese Academy of Sciences.
References
M.-S.P., W.-Z.W. and Y.-P.Z. designed research. M.-S.P., L.F., N.-N.S. and T.N. preformed the analysis. W.-Z.W, N.-N.S. and M.-S.P. constructed the website. L.F., N.-N.S. and M.-S.P. coded the software. M.-S.P. Y.-G.Y., R.W.M. and Y.-P.Z. wrote the manuscript.
Data Accessibility
The DomeTree is freely available on the web at http://www.dometree.org. The MitoToolPy and user manual are accessible on http://www.mitotool.org/mp.html.
The mtDNA sequence alignments were deposited in Dryad (doi:10.5061/dryad.cc5kn).




