cba-miR-222-3p involved in photoperiod-induced apoptosis in testes of striped hamsters by targeting TRAF7
Abstract
The role of miRNAs in the regulation of seasonal reproduction in rodents, particularly in relation to photoperiod changes, is still poorly understood. Previous studies on miRNA transcriptomes of striped hamster (Cricetulus barabensis) testes have indicated that the photoperiodism of testes, especially apoptosis, may be influenced by miRNAs. As a functional miRNA, cba-miR-222-3p in striped hamster testes exhibits suppression under a short photoperiod. To elucidate the potential role of testicular cba-miR-222-3p in the seasonal reproduction of striped hamsters, we exposed male striped hamsters to different photoperiods or injected miRNA agomir into the testes and observed the effects of these treatments, particularly some indicators related to apoptosis. The results showed that the levels of apoptosis in the testes increased in short daylength, accompanied by a significant decrease in cba-miR-222-3p expression and an increase in TRAF7 expression. Dual luciferase reporter assays verified the targeting relationship between cba-miR-222-3p and TRAF7 predicted by bioinformatics. In addition, the expression of TRAF7 decreased in the testes, which injected miRNA agomir, leading to inhibition of apoptosis, and the expression of key genes (MEKK3, p38, p53) in the downstream MAPK signaling pathway of TRAF7 was suppressed. These results suggest that short daylength induces testicular apoptosis in striped hamsters, and one possible mechanism is that the decreased expression of miR-222-3p in testes reduces the repression of TRAF7 translation, thereby activating the MAPK pathway and affecting the level of testicular apoptosis. These findings reveal the potential role of miR-222-3p in animal reproduction and provide new insights into the regulation of rodent populations.
INTRODUCTION
Animals inhabiting temperate zones exhibit a tendency to engage in breeding activities during specific seasons, while remaining reproductively inactive for the remainder of the year (Paniw et al. 2019). This adaptive behavior can be attributed to the evolutionary response of animals to the seasonal attributes of temperate regions (La et al. 2020). A seasonal breeder's reproduction is mainly modulated by photoperiod (Bohlen et al. 2018). A photoperiodic signal is transformed into a melatonin signal in the pineal gland (Schippers & Nichols 2014). Melatonin regulates the pulsed release of GnRH through Kisspeptin and then transmits the effect of light signals to the genital organs through the hypothalamic–pituitary–gonadal axis (Stevenson et al. 2012). In most rodent species, short winter photoperiods reduce testosterone levels, resulting in gonadal regression and decreased testosterone-dependent behaviors such as mating and aggression (Bedrosian et al. 2012). Although the inhibition of male hamster reproduction by short photoperiod has been fully confirmed, the molecular mechanism in the testes is not completely clear. Our previous research found that the striped hamster testes showed changes in endocrine, activation of apoptosis signal pathway, fluctuations in mitochondrial homeostasis, changes in miRNA expression profiles, and other phenomena during atrophy (Li et al. 2015; Zhao et al. 2022; Wang et al. 2023). However, the causes of these changes have not been fully revealed.
In recent years, research on the effects of miRNA on animal physiological processes has become a hotspot. A functional miRNA, miR-222-3p, has received much attention from researchers. During the healing process of rat fractures, miR-222-3p is believed to be associated with angiogenesis (Takahara et al. 2018). In obesity, upregulation of miR-222-3p may facilitate apoptosis in visceral adipose tissue (Wijayatunga et al. 2018). It has also been confirmed to exhibit age and gender differences in the physiological processes of the mouse nervous system (Rani et al. 2022). The above research indicates that miR-222-3p is widely expressed in the body and can actively participate in the physiological regulation process of rodents. Our previous study on miRNA expression profiles using miRNA-seq technology showed that the expression of cba-miR-222-3p of striped hamster testes decreased under a short photoperiod (Wang et al. 2023). Preliminary analysis of target genes indicated that cba-miR-222-3p may regulate the downstream mitogen-activated protein kinase (MAPK) signaling pathway by targeting TNF receptor-associated factor 7 (TRAF7). However, the underlying mechanism and phenotypic effects of cba-miR-222-3p in rodents still need further investigation.
TRAF7 is the last member of the tumor necrosis factor receptor (TNF-R)-associated factors (TRAFs) that have been identified. TRAF family was characterized as signaling adaptor molecules coupled to the cytoplasmic regions of receptors of the TNF-R superfamily (Zotti et al. 2012). TRAFs function as both scaffolds and enzymatic proteins for regulating the activity of MAPKs and nuclear factor-κB family transcription factors (NF-κBs) (Zotti et al. 2017). Multiple studies have confirmed that some members of this family and their upstream and downstream genes are regulated by miRNA (Bai et al. 2024; Selvakumar et al. 2024; Varshan et al. 2024). In the past few years, a series of evidence has been discovered that TRAF7 activates cell apoptosis, confirming the important role of TRAF7 in regulating physiological processes (Ding et al. 2018; Zou et al. 2023).
The striped hamster, Cricetulus barabensis, is one of the most destructive rodent pests in farmland and grassland of northern China, Mongolia, and Siberia (Xue et al. 2021). Therefore, the mechanism of population fluctuations in striped hamsters under photoperiod regulation has attracted the attention of researchers. As with other rodents that breed seasonally, striped hamsters display marked changes in reproductive physiology in response to different photoperiods (long daylength [LD], moderate daylength [MD], and short daylength [SD]) (Kelestimur et al. 2012), the most significant of which is testicular regression under SD.
The aim of this study is to explore the potential involvement and mechanisms through which miR-222-3p influences testicular physiology, especially the level of apoptosis, under different photoperiods. We initially assessed the level of apoptosis and the expression of cba-miR-222-3p in the testicles of striped hamsters exposed to different photoperiods. Subsequently, we employed bioinformatics methods to predict target genes, followed by dual luciferase experiments to validate the relationship between cba-miR-222-3p and its potential target, TRAF7. Finally, in vivo experiments were conducted by injecting miRNA agomir into the testicles of live striped hamsters to observe the potential impact of cba-miR-222-3p on the level of apoptosis in the testicular tissue. The result unveils a novel role of miR-222-3p in the reproductive regulation of seasonal reproduction in rodents. It also offers new insights into the control of rodent populations on miRNA.
MATERIALS AND METHODS
Ethical statement
All animal protocols were in accordance with Laboratory Animal Guidelines for the Ethical Review of Animal Welfare (GB/T 35892-2018) and were approved by the Biomedical Ethical Committee of Qufu Normal University (Permit Number: 2023121). This study was performed in compliance with all ARRIVE 2.0 guidelines.
Animal grouping and sample preparation
The striped hamsters used in this study were provided by the Experimental Animal Center of Qufu Normal University, and the original samples of its population were captured from farmland in Wucun Town, Qufu, China. Water and standard mouse chow were provided ad libitum during the experimental treatment. All hamsters were kept in animal isolation cages (NKsystem, Japan) with independent lighting and temperature control systems.
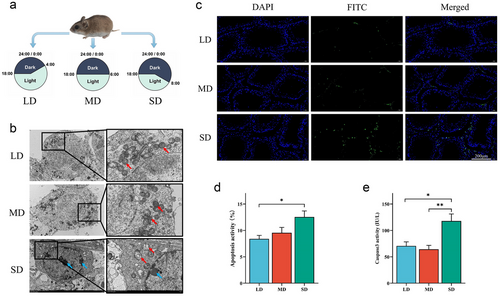
Photoperiod treatment experiment: Eighteen 6-month-old adult male hamsters (weighing 28 to 32 g) were evenly divided into three groups and subjected to different daylengths for 30 days. Detailed grouping and illumination time settings are shown in Fig. 1a. After treatment, all animals were sacrificed via CO2 asphyxiation between 10 and 11 p.m. All left testicles of each group were promptly collected and cut into two parts. One part was frozen in liquid nitrogen, then stored at −80 °C for real-time quantitative PCR (qRT-PCR), western blot analysis, and Caspase3 activity detection. The other four testicles in each group were immersed in 4% paraformaldehyde for tissue microarray and subsequent experiments (terminal deoxynucleotidyl transferase biotin-dUTP nick end-labeling [TUNEL], fluorescence in situ hybridization [FISH], and immunohistochemistry [IHC]). The remaining testicular tissues were immersed in glutaraldehyde for transmission electron microscopy.
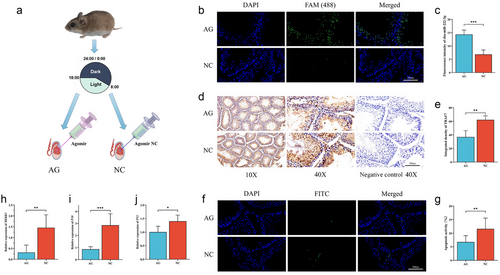
In vivo validation experiment: Another 12 hamsters treated in SD for 30 days were divided into two groups on average. One group was injected with cba-miR-222-3p agomir into the left testicle (AG), while the other group was injected with agomir naive control (NC); after injection, fed for another 5 days (Fig. 4a). The method for obtaining testicles was the same as above. The method for obtaining and dissecting the testicles was the same as described above. One part was frozen in liquid nitrogen and then stored at −80°C.
Tissue microarray
The tissue microarray (TMA) technique has established itself as a powerful approach for preparing and staining multiple tissue samples simultaneously and analyzing them simultaneously. A TMA is composed of many small tissue cores with a defined diameter that are fixed in a regular order on a single object carrier. First, the testicular tissues fixed with 4% paraformaldehyde were embedded in paraffin blocks. Sections of each tissue, with a thickness of 5 µm, were prepared and subjected to hematoxylin–eosin staining. A microscope was to observe and determine the target area. In the same area of the paraffin block, a hollow needle was used to obtain tissue cores as small as 2 mm in diameter. A precise array was then formed by transferring these tissue cores to a recipient paraffin block (Toberer et al. 2013). Then, 5–10 µm slices were cut from TMA using a slicer and transferred to poly L-lysine coated glass slides to prepare for subsequent experiments.
Transmission electron microscopy
The testes were investigated under a transmission electron microscopy (TEM, Hitachi, HT7800, Japan), as described previously (Wang et al. 2019b). The morphology of the nucleus and mitochondria is observed at a magnification of 10 000×.
Terminal deoxynucleotidyl transferase biotin-dUTP nick end-labeling (TUNEL) staining
Previously prepared TMA sections were taken and stained with a TUNEL staining kit (#MK1023, Boster, Wuhan, China). DAPI (#D1306, Sigma-Aldrich, France) staining was performed to count the number of nuclei. DNA breaks were determined using fluorescein isothiocyanate isomer. Sections were treated with DNase I for 10 min at room temperature before incubation with the TUNEL reaction mix. An upright fluorescence microscope (ECLIPSE CI, NIKON, Japan) was used to collect images, and an imaging system (DS-U3, NIKON, Japan) was used to scan and visualize the images. Each group had four individuals and three fields per individual were analyzed.
Caspase3 activity
Samples stored at −80°C were used to detect caspase-3 activity. For this, about 0.1 g of tissue was ground in 1 mL of PBS (0.01 mM, pH 7.2) on ice and after 15 min incubation on ice, centrifuged at 4°C for 10–15 min to obtain the cell lysates by taking the supernatant. Following the manufacturer's instructions, caspase-3 activity was measured in lysates using the Caspase-3 Activity Kit (BC3830, Solarbio, China).
Fluorescence in situ hybridization
The fluorescence in situ hybridization (FISH) method was adapted from Fay and John (Wang et al. 2012). Briefly, previously prepared TMA sections were deparaffinized in xylene, followed by dehydration in an ethanol series. After incubation in citrate buffer (10 nmol L−1, pH 6) for 10–15 min, the TMA sections were rinsed in deionized water and immediately treated with 20 µg mL−1 protease K at 37°C for 30 min. Subsequent hybridization (the excitation wavelength of FAM(488) green light was 465–495 nm), washing, and DAPI staining were performed according to the instructions of the Fluorescence in Situ Hybridization Kit for RNA (R0306S, Beyotime, China). Images were collected using a fluorescence microscope (ECLIPSE CI, NIKON, Japan), and images were scanned and visualized using an imaging system (DS-U3, NIKON, Japan). Each group had four individuals, and three fields per individual were analyzed.
Real-time quantitative PCR
Total RNA extraction, cDNA synthesis, and qRT-PCR were performed as described (Wang et al. 2023). Briefly, total RNA was extracted from testicular tissue using TRIZOL method. The extraction process strictly follows the instructions of RNAiso Plus Kit (TaKaRa, Dalian, China). The cDNA of mRNA and miRNA were synthesized by Evo M-MLV RT Premix for qPCR (AG, Hunan, China) and miRNA 1st strand cDNA synthesis kit (contains universal primer for qRT-PCR) (AG, Hunan, China), respectively. qRT-PCR was performed using a SYBR Green Premix Pro Taq HS qPCR Kit II (AG, Hunan, China). The reference genes were U6 snRNA for miRNA quantification and β-actin for mRNA quantification. The results were examined using the 2−ΔΔct method. The primers used for qRT-PCR are shown in Table 1.
| Name | Sequence (5′ to 3′) | Annealing temperature (°C) |
|---|---|---|
| U6 | GCTTCACGAATTTGCGTGTC | 60 |
| cba-miR-222-3p | AGCTACATCTGGCTACTGGGTCTCT | 60 |
| MEKK3 | F: GCTCTCCTCCACCTGGTTACG | 59 |
| R: ATACACTGCTCGCTGGTCTCTG | ||
| P38 | F: GCTGTGACCTTCTTGGCTAACC | 59 |
| R: GAACCTGATCCGTGGAAGTGTAG | ||
| P53 | F: AAGTTGTAAGACGCTGCCCTCAC | 61 |
| R: ACTGTGCCGAAATGTTTGCTTGTC | ||
| β-actin | F: GAGACCTTCAACACCCCAGC | 59-61 |
| R: ATGTCACGCACGATTTCCC |
Prediction and functional analysis of miRNA and target genes
The target gene predictions were performed by three software programs: Miranda 3.3a, RNAhybrid 2.1, and TargetScan 7.0. Predicted miRNA target genes were selected from intersections of the results. Then, a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed on the target genes.
Immunohistochemistry
IHC was performed as described (Blakely et al. 2015). Briefly, previously prepared TMA sections were stained with the TRAF7 antibody (11780-1-AP, Proteintech, China, RRID:AB_2877793) per the manufacturer's instructions. A microscope (E100, NIKON, Japan) was used to collect images and an imaging system (DS-U3, NIKON, Japan) was used to scan and visualize the images. Positively stained cells were measured using three fields of view per section. The mean and standard error of that mean were calculated. Each group had four individuals and three fields per individual were analyzed.
Dual-luciferase reporter assay
The binding site between TRAF7 3′-untranslated region (3′UTRs) fragment and miR-222-3p were predicted using starBase (http://starbase.sysu.edu.cn/). To validate the microRNA-binding sequence of miR-222-3p, fragments containing wild-type (WT) or mutated-type (MT) TRAF7 3′UTRs of the predicted binding site were cloned into the PmirGLO vector, which was constructed by Sangon Biotech, Inc. The miR-222-3p mimic and NC mimic were also synthesized by Sangon Biotech, Inc. 293 cells were co-transfected with miR-222-3p mimic or NC, and pmirGLO-TRAF7-3′UTR WT or pmirGLO-TRAF7-3′UTR MT, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the instruction of the manufacturer. Forty-eight hours post-transfection, the firefly and Renilla luciferase activities were determined by Dual-Luciferase reporter assay (Promega, USA). Briefly, 100 µL Passive Lysis Buffer was added to the 96-well plate and centrifuged at 1200 × g at 4°C for 10 min, and then 100 µL Luciferase Assay Reagent II and 100 µL cell lysate were added in sequence and mixed by pipetting two to three times. 100 µL STOP & GLO® reagent (Luciferase Assay Reagent; Promega Corporation) was added and mixed two to three times to record the Renilla luciferase value, which was the reporter gene luminescence value.
Western blot
Homogenization of tissue samples was performed in RIPA lysis buffer. PierceTM BCA Protein Quantification Kit (#53225, Thermo Fisher, USA) was used to determine the amount of soluble protein. The samples were then adjusted to a final protein concentration of 3 µg µL−1 with SDS buffer and homogenizing buffer. Equal amounts of protein from each sample (20 µL) were loaded onto 10% polyacrylamide gels containing 0.5% trichloroethanol and run through SDS–PAGE. After performing electrophoresis, the proteins were transferred to PVDF membranes, which were then incubated with TRAF7 antibodies (11780-1-AP, Proteintech, China, RRID:AB_2877793) at a 1:1000 dilution and β-actin antibodies (20536-1-AP, Proteintech, China, RRID:AB_10700003) at a 1:1000 dilution, and subsequently incubated with IRDye 800 CW goat anti-rabbit secondary antibodies (31 460, Thermo Fisher, USA). The membranes were finally visualized by an Odyssey scanner (Bio-Rad, USA). The blots were quantified using ImageJ software.
In vivo injection
miRNA agomirs are specially labeled and chemically modified small RNAs that regulate the biological function of target genes by mimicking the entry of endogenous miRNA into the miRiSC complex. The cba-miR-222-3p agomir (AG) and agomir negative control (NC) used in this study were designed and synthesized by Sangon Biotech Inc. The drug was purified by high-performance liquid chromatography method. 1 nmol of drug powder requires the addition of 5 µL of DEPC water. The final concentration of the solution is 200 pmol µL−1. Agomir and agomir NC were injected into the left testes using a microsyringe. Each left testis was injected a volume of 10 µL.
Statistical analyses
Shapiro–Wilk and Levene tests were applied to test for normality and homogeneity of variance, respectively. One-way ANOVA was used to compare the differences between groups. Statistical differences between groups were tested using t-test. The differences were considered significant at P < 0.05. Data were expressed as means ± standard deviation (mean ± SD). All statistical analyses were performed using SPSS 26.0.
RESULTS
Apoptosis of striped hamster testes under different photoperiods
Three methods have been used to detect apoptosis in the testicles of striped hamsters. Using transmission electron microscopy, early apoptotic phenomena in spermatocytes of SD can be observed, such as mitochondrial vacuolization and nuclear edge pyknosis (Fig. 1b). TUNEL staining directly demonstrated apoptosis in the cells. Green fluorescence indicates fragmentation of DNA. With the decrease of photoperiod, the green fluorescence in the testes significantly increased, indicating an increase in the level of apoptosis (Fig. 1c; Table S1, Supporting Information). The location of enhanced fluorescence suggested that apoptosis occurred mainly in the interstitial cells, and there was also an increase in the proximity of the germinal epithelium to the seminiferous tubule wall. The statistical result of TUNEL detection showed that the proportion of apoptotic-activated cells in the SD group was significantly higher than that in the LD group (P < 0.05; Fig. 1d). To further confirm the increased level of apoptosis in the testes of SD, the activity of Caspase3 was detected by ELISA. The activity of caspase3 in SD was significantly higher than that in LD (P < 0.05) and MD (P < 0.01) (Fig. 1e; Table S2, Supporting Information). Although the three methods used for detecting apoptosis levels were not entirely consistent, there was a collective affirmation of the increase in apoptosis levels in the SD group.
Expression of cba-miR-222-3p in testes of striped hamsters under different photoperiods
The expression of cba-miR-222-3p in testes was detected by FISH (TMA) and qRT-PCR method. In the FISH images, the green fluorescence area indicated the expression of cba-miR-222-3p (Fig. 2a). The intensity of green fluorescence decreased as the photoperiod shortened. In the LD and MD groups, green fluorescence was observed in various types of cells, whereas in the SD group, the fluorescence was primarily concentrated in spermatocytes, with weak intensity. Statistical results of fluorescence intensity indicated that the SD group was significantly lower than the other two groups (P < 0.01), consistent with the results shown in the images (Fig. 2b; Table S3, Supporting Information). The qRT-PCR result also showed that the LD group was significantly higher than the SD group (P < 0.05) (Fig. 2c; Table S6, Supporting Information). They collectively confirmed that cba-miR-222-3p was down-regulated in the testes of striped hamsters under short photoperiod.

Target genes and functional prediction of cba-miR-222-3p
Three prediction tools (RNAhybrid/miranda/targetscan) were used to predict the target genes of cba-miR-222-3p. Due to different principles of prediction, the results obtained by the three tools are also different. The intersection of three predicted results will be considered a more reliable result. In Fig. 2d, 316 targets were predicted by three tools. Functional analysis of these targets through KEGG enrichment can provide valuable insights into the biological pathways and processes that are associated with these targets. The results showed that these target genes were mainly enriched in Proteoglycans in cancer, Tight junction, Focal adhesion, Calcium signaling pathway, Axon guidance, Salmonella infection, and MAPK signaling pathway (Fig. 2e). By analyzing the functions of all targets and the stability of their binding with cba miR-222-3p individually, we speculated that the binding between cba miR-222-3p and TRAF7 is relatively strong, and TRAF7 has a robust correlation with apoptosis.
cba-miR-222-3p directly targets TRAF7
To further evaluate whether cba-miR-222-3p is functional in targeting TRAF7 3′UTR, we performed the Dual-Luciferase Reporter Assay. As shown in Fig. 3a, there is a binding site containing eight bases on the 3′UTR of TRAF7 mRNA, and we mutated the binding site. The WT and MT 3′UTR (mutated at positions 271–278) sequences of TRAF7 were cloned into the pmirGLO vector. miR‑222-3p mimic and mimic negative control were co-transfected into cells with the pmirGLO-TRAF7-3′UTR WT and pmirGLO-TRAF7-3′UTR MT plasmid. The dual luciferase detection system then detected luciferase activity. The result showed that when miR-222-3p mimic was co-transfected with WT, the luciferase activity was significantly lower than when co-transfected with MT. However, when miR-222-3p mimic is replaced with NC mimic, luciferase activity remained at a higher level regardless of the presence of WT or MT (Fig. 3b; Table S5, Supporting Information). This indicated that cba-miR-222-3p performs post-transcriptional regulation by targeting the 271–278 positions of 3′UTR of TRAF7.
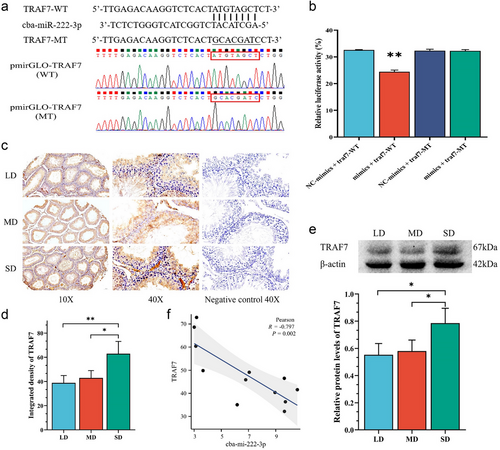
Expression of TRAF7 in striped hamster testes under different photoperiods
The expression of TRAF7 in striped hamster testes was detected by IHC (TMA) and western blot. The result of IHC showed that, compared to the LD and MD groups, the SD group exhibited darker staining, indicating a stronger positive reaction, that is, higher TRAF7 expression (Fig. 3c). By evaluating the integrated density of IHC photos, it was found that the SD group was significantly higher than the MD group (P < 0.05) and extremely significantly higher than the LD group (P < 0.01; Fig. 3d; Table S4, Supporting Information). The trend of the Western blotting result is similar to the IHC result and also shows an increase in TRAF7 expression in the SD group (Fig. 3e). Correlation analysis showed a significant correlation between cba-miR-222-3p and the expression level of TRAF7 (R = −0.797, P = 0.002; Fig. 3f)
Detection of TRAF7 and apoptosis in testicles after injection of agomir
To further investigate the role of cba-miR-222-3p in the testes of living hamsters, cba-miR-222-3p agomir and its negative control were synthesized and injected into the testes of striped hamsters. Using FISH (TMA) to detect the expression of miR-222-3p, it can be observed from the images that the AG group has more green fluorescence than the NC group (Fig. 4b; Table S3, Supporting Information). The intensity of green fluorescence in the AG group was significantly higher than that in the NC group (P < 0.001; Fig. 4c). This indicated that agomir was successfully injected and stably expressed in the testes of hamsters. At the same time, TRAF7 was also detected in testicular tissue. The results of IHC (TMA) showed a significant decrease in TRAF7 expression in the AG group (P < 0.01; Fig. 4d,e; Table S4, Supporting Information); TUNEL (TMA) detection results showed that the AG group had less green fluorescence (Fig. 4f; Table S1, Supporting Information). This means that the AG group has less gene fragmentation. Statistics showed that the percentage of positive cells in the AG group was significantly lower than that in the NC group (P < 0.01; Fig. 4g). This indicated a decrease in apoptosis in the AG group. To confirm the pathways that affect apoptosis, the expression of key genes on the downstream MAPK pathway was measured. MEKK3 is thought to interact with TRAF7, and the expression of MEKK3 in the AG group was notably lower than that in the NC group (P < 0.01; Fig. 4h). Additionally, the expression of p38 in the AG group was significantly lower than in the NC group (P < 0.001; Fig. 4i), and the expression of p53 was also notably lower than in the NC group (P < 0.05; Fig. 4j; Table S7, Supporting Information). These findings indicated that the decreased apoptosis in the AG group may be attributed to the inhibition of the MAPK signaling pathway.
DISCUSSION
In this study, we investigated the increase in apoptosis levels in the testes of striped hamsters under short photoperiod and one possible miRNA mechanism underlying this phenomenon. The study of photoperiodic regulation of the testes is an area of intense interest because of its significance for understanding the mechanisms of circadian clock and reproductive regulation (Wang et al. 2019a). At present, many studies have explored the mechanism of photoperiod on the development and function of testes and its application in production and life, such as the use of photoperiod to regulate the growth and reproduction of livestock and poultry (Kim et al. 2022; Mandal et al. 2022; Stewart & Marshall 2022). However, there are few studies on rodents. In this study, we investigated the molecular mechanism of photoperiod regulation in the testes of striped hamsters by setting photoperiod conditions consistent with their habitats. This study unveils a novel role of miR-222-3p in the reproductive regulation of seasonal reproduction in rodents. It also offers new insights for ecological research on miRNA.
Photoperiod affects apoptosis in the testis of striped hamsters
Mammals use photoperiodic changes to adapt to the alternating between breeding and non-breeding seasons (Guh et al. 2019). Seasonal breeding mammals undergo a series of physiological changes during the non-breeding season. These changes include a decrease in body temperature, a decrease in metabolic rate, a decrease in energy demand, and atrophy of the gonads (Zhang et al. 2017). This allows mammals to better adapt to changes in their environment and survive harsh conditions. The atrophy of the testes in mammals is due to the regulatory role of the endocrine system (Tomihara et al. 2022). During the breeding season, the testicles produce large amounts of hormones, such as testosterone, to promote sperm production. However, in the non-breeding season, due to changes in the external environment and the regulation of the biological clock, the endocrine system will change, resulting in reduced testosterone secretion. This reduction leads to apoptosis of testicular cells and atrophy of testicular tissue (Xi et al. 2021; Valentini et al. 2022).
In this study, the increased apoptosis of testicular tissue in the SD group was confirmed by TUNEL detection, electron microscopy observation, and caspase3 activity detection. The results are consistent with the researchers' work on the Syrian hamster and Iberian mole (Dadhich et al. 2010; Martínez-Hernández et al. 2020). Although there were differences in the duration and method of treating hamsters compared to other studies, our results confirmed two points. First, the level of apoptosis in the testes of striped hamsters was indeed regulated by photoperiod. Second, we identified a time scale, within 30 days, in which the influence of the photoperiod could significantly manifest in the testes. Based on the studies in other species (Sun et al. 2020; Beltrán-Frutos et al. 2022), we can roughly infer that the period of 20–30 days may be a critical initial or early stage for testicular response to photoperiodic regulation. This provided a very important clue for research in animal physiology and reproduction.
Apoptosis is a normal cell death process that plays an important role in maintaining the balance of tissues and organs (Soriano & Scorrano 2011). During the non-breeding season, cells within the testes undergo apoptosis, a regulated cell death process that drives cell death and resorption within the testes, resulting in testicular atrophy. Our findings support this perspective. When exposed to simulated non-breeding season photoperiods, the testes of striped hamsters undergo increased apoptosis and reduced volume. Considering that under the conditions of SD, male hamsters still have a testis coefficient (testis weight/body weight) far higher than most other animals. Therefore, we do not believe that male hamsters completely lose their reproductive ability during non-breeding seasons but rather reduce the energy allocation for reproduction, which may be a more prudent survival strategy (Ashley et al. 2012).
cba-miR-222-3p mediates photoperiod-induced apoptosis in the testes by targeting TRAF7
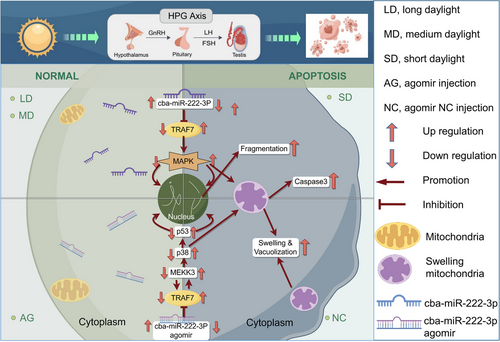
The expression of cba-miR-222-3p was found to be decreased in the SD group. This is consistent with the result of previous studies on miRNA transcriptome (Wang et al. 2023). Together, they confirmed that the SD condition had an inhibitory effect on the expression of cba-miR-222-3p. Based on bioinformatics methods, 316 potential target genes were predicted by three prediction tools. Among these target genes, TRAF7 is thought to play an important role in the process of apoptosis by regulating the MAPK and NF-κB signaling pathways (Zotti et al. 2017; You et al. 2023). The direct targeting relationship between cba-miR-222-3p and TRAF7 was confirmed by dual luciferase assay. The regulation of TRAF7 by cba-miR-222-3p was also demonstrated by the in vivo injection of agomir into striped hamsters. After agomir injection, TRAF7 expression was decreased as well as apoptotic activity measured by TUNEL. This further demonstrates the inhibitory effect of cba-miR-222-3p on apoptosis in the hamster testes. After the injection of agomir, qRT-PCR experiments confirmed significant changes in the expression of some key genes (MEKK3, p38, p53) in the MAPK signaling pathway. Therefore, it is inferred that the cba-miR-222-3p/TRAF7 axis affects cell apoptosis through the MAPK signaling pathway.
The FISH images revealed that the most intense green fluorescence was observed in spermatogonia, indicating a high specific expression of cba-miR-222-3p in spermatogonia. This characteristic was observed in all three groups, suggesting that cba-miR-222-3p may play a role in the differentiation of spermatogonia. The decreased expression of cba-miR-222-3p in the SD group may lead to reduced differentiation of spermatogonia, consequently lowering the rate of sperm production (Dong et al. 2014). This may be one of the reasons why striped hamsters are inhibited in their reproductive activities during non-breeding seasons.
The role of miRNA in the post-transcriptional regulation of genes has been recognized by researchers (Dvinge et al. 2013; Kp et al. 2024). However, because the regulatory network of miRNAs is very complex, miRNA can regulate multiple target genes at the same time, and a target gene can also be regulated by multiple miRNAs. In addition, miRNAs can be adsorbed by lncRNAs and circRNAs (Cesana et al. 2011; Slack & Chinnaiyan 2019). As a result, the expression of miRNA in the same cell is challenging to exhibit significant differences under non-pathological conditions, and the regulation of miRNA on gene expression is relatively weak, that is, phenotypic effects may be more subtle when individual targets are gained or lost (Mendell & Olson 2012). This complexity makes it more difficult to study miRNA function and mechanism. This is even more evident in studies using live animals as experimental materials. Similar difficulties were encountered when searching for potential functional miRNAs based on small RNA transcriptome data. Among them, cba-miR-222-3p was successfully discovered and verified by dual luciferase assay and in vivo experiments. Due to the lack of relevant miRNA research, the proportion of the anti-apoptotic effect of cba-miR-222-3p cannot be accurately evaluated. However, with the gradual increase of experimental data, the effect of cba-miR-222-3p will be more reasonably evaluated.
In this study, we demonstrated for the first time that cba-miR-222-3p could target the 3′UTR of TRAF7 to reduce the expression of TRAF7 protein in the testes of striped hamsters. They have eight consecutive pairs of bases that can complement each other and form a relatively stable binding. Data from public databases (miRBase https://www.mirbase.org/) showed that miR-222-3p were highly conserved among different species. Interestingly, the TRAF7 3′UTR sequence was poorly conserved, and our validated target region was only found in some rodents. Although more data and studies are needed, it can be preliminarily inferred that the effects of the cba-miR-222-3p/TRAF7 axis are different among species. Although this is of minor relevance to this study, it provides species-specific targets for drug development based on cba-miR-222-3p and the control of the rodent population. Therefore, it is of significant importance.
However, there are still some deficiencies in our research. For example, fewer animal samples were used in this study due to difficulties in obtaining animal samples. In addition, although tissue microarray technology allows all tissues to be under absolutely the same conditions during the experiment, it is also inevitable that each tissue will have only a small field of view.
In summary, we studied the induction of testicular apoptosis in striped hamsters by exposing male striped hamsters to different photoperiods, and further explored one possible miRNA mechanism and its downstream signaling pathway. The shortened photoperiod reduces cba-miR-222-3p, leading to decreased inhibition of TRAF7 translation and increased expression of TRAF7 protein. Additionally, cba-miR-222-3p/TRAF7 axis activates the downstream MAPK signaling pathway, promoting cell apoptosis (Fig. 5). This research uncovers the potential role of miR-222-3p in animal reproduction and offers new insights for rodent population regulation.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Nos. 32072436, 31770455, 31972283, and 31800308).
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are included in the supporting information.




