Intraspecific diversity of Meriones persicus (Rodentia; Gerbillinae), the main plague reservoir in Iran, and its connection to enzootic plague in Iran
Abstract
Plague, a lethal zoonotic disease, primarily circulates within rodent populations and their fleas. In Iran, the widely distributed jird, Meriones persicus, serves as the principal reservoir for plague, with a belief in the existence of five out of its six recognized subspecies within the country. However, these subspecies are classified into four mitochondrial cytochrome b sub-lineages (IA, IB, IIA, IIB). This discrepancy, combined with the presence of an unnamed sub-lineage in central Iran awaiting taxonomic clarification, has left intraspecific taxonomy unsettled and obscured the true alignment between mtDNA sub-lineages and nominal subspecies. In this study, we investigated the intraspecific variation in the cytb gene across populations sampled throughout Iran, focusing on underexplored regions between the Zagros and Alborz Mountains and central Iran. While our genetic data generally support reported subspecies validity in Iran, we raise questions about M. p. baptistae, emphasizing the need for further data from its type territory in Pakistan. Two main lineages of M. persicus (I and II) exhibit geographical isolation, with limited overlap in the central Zagros Mts., where three subspecies (M. p. ambrosius, M. p. rossicus, and M. p. persicus) coexist. Superimposing infected rodents' geographic coordinates onto updated sub-lineages' distribution revealed a potential association between sub-lineage IA (M. p. rossicus) and all enzootic plague cases from 1946 to 2023. M. persicus rossicus extends into the Caucasus (where plague infections are common), Eastern Turkey, and Iraq. Consequently, interpreting this finding in the context of plague surveillance in Iran and neighboring areas requires caution.
INTRODUCTION
Rodents are key components of the majority of terrestrial ecosystems (Wilson et al. 2017) and also a source of emerging infectious diseases (EIDs) (Han et al. 2015; Rabiee et al. 2018; White & Razgour 2020). They are of utmost significance in plague dynamics, one of the deadliest bacterial pathogens, which still is a global concern. The EIDs’ epidemiology is associated with their reservoirs, which vary from one region to another (Bonvicino et al. 2015; Mahmoudi et al. 2021). Because of a species-specific role in the EIDs' dynamics (Eisen & Gage 2009), taxonomic information on rodent hosts is crucial for any surveillance systems (Bickford et al. 2007). The history of plague outbreaks in Iran has been well-documented by Shahraki et al. (2016), who recorded over 40 outbreaks between 1829 and 1966, resulting in numerous fatalities. The origin of the disease in Iran—whether introduced through trade, military campaigns, or endemic natural foci with continuous Yersinia pestis circulation—remains uncertain. The investigation teams of the Pasteur Institute of Iran identified the enzootic cycle of Y. pestis among rodents and its association with sporadic human outbreaks in western Iran during the 1940s to 1970s, which led to 156 deaths. This confirmed the endemic nature of plague in the area (Baltazard et al. 1952, 1960), and subsequent studies demonstrated that the infection is endemic and not introduced externally (Baltazard et al. 1963, 1964; Esmaeilie et al. 2013, 2023). Among the four jird species (genus Meriones) suspected as primary plague reservoirs in the Iranian focus, Meriones persicus and M. libycus display higher resistance to plague, while M. vinogradovi and M. tristrami exhibit greater susceptibility and mortality rates (Baltazard et al. 1963).
M. persicus, owing to its prevalence and high population density, is recognized as the principal species responsible for maintaining the plague in western Iran. Notably, among the 127 nonhuman mammals identified with plague infections in Iran over 77 years, 88 cases were attributed to M. persicus (Baltazard et al. 1952, 1960; Karimi 1980; Esmaeili et al. 2013, 2023).
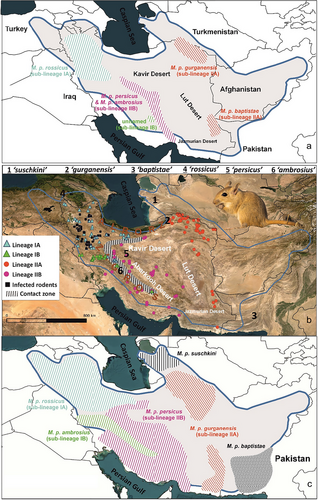
M. persicus (Gerbillinae; Muridae), like other gerbils, inhabits arid and semi-arid ecosystems including grasslands, foothills, cultivated areas, and rocky hillsides (Pavlinov et al. 1990). Its distribution covers the Iranian plateau, excluding the Caspian Sea shore. It extends from eastern Turkey, the trans-Caucasus, and Iraq in the west to Pakistan, Afghanistan, and Turkmenistan in the east (Fig. 1b). Denys et al. (2017) acknowledged six subspecies, namely M. p. persicus (Blanford, 1875), M. p. rossicus Heptner, 1931, M. p. ambrosius Thomas, 1920, M. p. gurganensis Goodwin, 1939, M. p. baptistae (Thomas, 1920), and M. p. suschkini (Kashkarov, 1925). All of these subspecies are believed to be present in Iran, except for M. p. suschkini, which was described from Les Arshevi within the Balkan Mts. in western Turkmenistan (Ellerman & Morrison-Scott 1951). Recent mitochondrial cytb gene analysis identified two main lineages subdivided into four phylogenetic sub-lineages in Iran (IA, IB, IIA, and IIB; see Fig. 1a following Dianat et al. 2017). These sub-lineages did not align neatly with traditional subspecies (Ellerman & Morrison-Scott 1951; Denys et al. 2017). Sub-lineage IA (M. p. rossicus, cf Dianat et al. 2017) occurs in Armenia, eastern Turkey, Iraq, and northwestern Iran; sub-lineage IB (an unnamed lineage, cf. Dianat et al. 2017) occupies Shirkooh Mts. in central Iran (Yazd Province); sub-lineage IIA (encompassing M. p. gurganensis and M. p. baptistae, cf. Dianat et al. 2017) is in eastern Iran spanning from Khorasan in the northeast to the Sistan and Baluchestan province in the southeast; and sub-lineage IIB (encompassing M. p. persicus and M. p. ambrosius cf. Dianat et al. 2017) is composed of specimens from the Qohrud Mts. and southern Zagros Mts. (Fig. 1a) (Dianat et al. 2017).
Recent advances in rodent taxonomy have led to significant changes in the list of plague reservoir species (Bonvicino et al. 2015; Mahmoudi et al. 2021). Our contemporary comprehension of biodiversity, evolution, taxonomy, and the delineation of hosts, vectors, pathogens, and their interactions in the epidemiology of EIDs has significantly profited from enhanced access to genetic material (Bickford et al. 2007; Webster et al. 2016). Western Iran, particularly the regions between the Zagros Mts. and the Alborz Mts., houses a principal plague focus in the Middle East (Baltazard et al. 1964), concurrently emerging as a hotspot of rodent diversity (Yousefi et al. 2022, 2023). In this study, we adopted an integrative approach to explore the intraspecific variation and taxonomy of M. persicus. Specimens collected from diverse locations in Iran underwent morphological comparisons based on four standard external measurements. Furthermore, molecular assessments were carried out utilizing the mitochondrial cytochrome b (cytb) gene, acknowledged as a reliable marker for intraspecific geographic variations in mammals (Nicolas et al. 2012; Galimberti et al. 2015). Subsequently, we superimposed a georeferenced map of the enzootic plague (Baltazard et al. 1952, 1960; Karimi 1980; Esmaeili et al. 2013, 2023) onto the spatial distribution of M. persicus phylogenetic sub-lineages, seeking potential correlations. Our study unveiled taxonomy as a meaningful tool for classifying infraspecific variation within M. persicus. Notably, the observed connection between the enzootic plague range and the geographic distribution of a specific subspecies (M. p. rossicus) underscores the necessity of accounting for infraspecific reservoir structure concerning the epidemiology of rodent-borne infectious diseases, warranting deeper consideration.
MATERIALS AND METHODS
Sample collection and morphological assessment
Our objective was to cover the entire distribution of M. persicus in Iran, but focusing particularly on the regions that have received limited research attention, as highlighted by Dianat et al. (2017), specifically, the areas along the Zagros and Alborz Mts. and central massif (Fig. 1b). All specimens are preserved as standard museum skins/and or skulls and deposited at the Museum of Vertebrate Zoology, Berkeley, USA, Mohammad Hanifi's Health Museum, Pasteur Institute of Iran, Natural History Museum of Urmia University, Iran, and Zoological Museum of Ferdowsi University of Mashhad, Iran (see specimens detail in Table S1, Supporting Information). Specimens were morphologically identified at the species level using established keys (Kryštufek & Vohralík 2009; Darvish et al. 2014). Four external measurements including head and body length (HBL), tail length (TL), hind-foot length, and ear length were scored using a ruler to the 0.1 accuracy.
The current study used material deposited in collections, hence no collecting permits were required.
DNA extraction, PCR amplification, and DNA sequencing
DNA extraction was performed on 74 alcohol-preserved spleen or muscle specimens utilizing the DNeasy tissue kit (Qiagen, Germany). Amplification of the complete mitochondrial cytochrome b (cytb) gene was carried out using primers L14727-SP–H-15 915-SP (Jaarola & Searle 2002) and L7–H6 (Montgelard et al. 2002). The PCR protocols adhered to the methodology outlined by Jaarola and Searle (2002). Subsequently, PCR products were purified with the QIAquick PCR purification Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocol. For commercial sequencing, dye-labeled dideoxy terminator cycle sequencing with Big Dye V.3.1 (Applied Biosystems, Inc., Macrogen Company, South Korea) was employed.
Phylogenetic inference
The 74 newly acquired sequences were edited, aligned using CodonCode Aligner software (CodonCode Corp.), and compared to available M. persicus cytb sequences (n = 71) from GenBank (Table S1, Supporting Information). jModeltest (Posada 2008) was used to determine the appropriate substitution model based on the Akaike Information Criterion (AIC). The maximum likelihood (ML) phylogenetic tree was constructed via the RAxML web server (https://cme.h-its.org/exelixis//web/software/raxml/), generating bootstrap values (ML-BS) from 1000 replicates (Stamatakis 2014). Bayesian Inference (BI) were generated with MrBayes v3.2 (Ronquist et al. 2012) using the Markov chain Monte Carlo method. Two independent runs were initiated from random trees, each running for 2 × 106 generations and sampled every 1000 steps with a relative burn-in of 25%. Branch support was assessed by Bayesian posterior probabilities (BPP) in the BI tree and Bootstrap in the ML tree (BP) assuming >0.95 BPP and >80 BP as good supports. Phylogenetic trees were rooted using midpoint rooting algorithm. The phylogenetic relationships between haplotypes were also determined by constructing a network using the median-joining method, as provided in NETWORK v4.500 (Bandelt et al. 1999).
Genetic variability and demographic tests
We employed the Kimura-2 parameter model, using pairwise deletion algorithm, to compute genetic divergence among mtDNA sub-lineages using MEGA v7.0 (Kumar et al. 2016). DnaSP v5.10 (Librado & Rozas 2009) estimated the genetic structure of molecular sub-lineages, encompassing polymorphic site count, average nucleotide differences, nucleotide diversity, and haplotype diversity. For investigating sub-lineage demographic history, we employed mismatch distribution analysis under pure demographic expansion model, Tajima's D (Tajima 1989), and Fu's Fs tests (Fu 1997), influential neutrality tests that can elucidate population growth, decline, and stability. ARLEQUIN v3.5 (Excoffier & Lischer 2010) was utilized for these analyses, with 1000 coalescent simulations to validate significant values (P < 0.02 and P < 0.05 for Fs and D statistics, respectively).
Correlation between enzootic plague and M. persicus mtDNA sub-lineages
Georeferenced locations of Yersinia pestis-infected rodents, the causative agent of plague, were sourced from published references (Baltazard et al. 1952, 1960; Mostafavi et al. 2017, 2018; Esmaeilie et al. 2013, 2023) and unpublished field reports from Pasteur Institute of Iran spanning 77 years. The integration of enzootic plague projections and the distribution of phylogenetic sub-lineages on a georeferenced map was conducted using QGIS 3.22.5 (https://qgis.org).
RESULT
External morphology and measurements
Persian jird can be distinguished from other Meriones species in Iran by having completely naked palms and soles, a long tail covered in short hairs that end in a large terminal pencil, a pure white belly, and relatively large ears (>19 mm) (Fig. S1, Supporting Information). Cranially, it possesses traits such as a relatively short braincase and an elongated rostrum. External measurements of the four mtDNA sub-lineages/nominal subspecies (M. p. rossicus, M. p. ambrosius, M. p. persicus, and M. p. gurganensis) in Iran are outlined in Table 1. Although the tail is relatively longer in lineage I and head and body is larger in lineage II, there was no statistically significant difference between lineages (P > 0.05).
| Subspecies | mtDNA sub-lineage | HBL | TL | HFL | EL | HBL/TL |
|---|---|---|---|---|---|---|
| M. p. rossicus (n = 21) | IA | 147.57 ± 15.39 | 164.13 ± 17.25 | 37.83 ± 3.63 | 22.39 ± 3.09 | 89% |
| M. p. ambrosius (n = 79) | IB | 141.69 ± 15.58 | 172.48 ± 19.79 | 38.85 ± 2.62 | 23.22 ± 2.73 | 81% |
| M. p. gurganensis (n = 30) | IIA | 152.80 ± 15.26 | 152.26 ± 21.82 | 36.85 ± 2.16 | 22.68 ± 3.72 | 90% |
| M. p. persicus (n = 15) | IIB | 150.15 ± 13.59 | 158.80 ± 16.02 | 38.26 ± 8.54 | 23.37 ± 3.80 | 94% |
- HBL, head and body length; TL, tail length; HFL, hind-foot length; EL, ear length.
Phylogeny
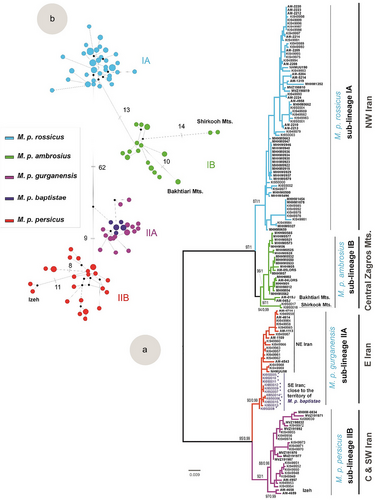
The selected model based on the AIC (GTR + I + G, I = 0.5848, G = 0.8937) was applied to construct phylogenetic relationships between M. persicus populations. Both ML and BI trees shared the same branching structure (BP > 85, BPP > 0.99); therefore, only ML tree is shown (Fig. 2a). Two main lineages were present within M. persicus (I, II) with subsequent divisions into four sub-lineages (IA, IB, IIA, and IIB). Great majority of Iran was populated by lineage II (hereafter “persicus” group) subdivided into sub-lineages IIA (stretching from northeastern Iran in Khorasan to southeastern in Sistan and Baluchistan) and sub-lineage IIB (encompassing the Zagros Mts. and an isolate in the Alborz Mts.). Meanwhile, western Iran was predominantly inhabited by lineage I (hereafter “ambrosius” group) further dividing into sub-lineages IA (northwestern Iran) and IB (central Zagros Mts. in Lorestan and Chaharmahal and Bakhtiari provinces, along with an isolate in Shrikoh Mts. in Yazd Province). Lineages I and II exhibited primarily allopatric distributions across the eastern and western Iranian plateau, with a slight parapatric overlap around the Zagros Mts. (Fig. 1b). Notably, while lineage II primarily occupied eastern Iran, it also extended into the central and southwestern parts of the country, including the Zagros Mts., the region that is populated by three parapatric sub-lineages (IA, IB, and IIB). Specifically, populations from the northern Zagros Mts. (West Azerbaijan, East Azerbaijan, Hamadan, Kermanshah, and Kurdistan provinces), populations from the central Zagros Mts. (eastern Lorestan and northwestern Isfahan provinces), as well as those from the southern slope of the Alborz Mts. (Qazvin, Zanjan, and Tehran provinces including Lar and Khojir National parks) were affiliated with sub-lineage IA. The majority of populations from the central Zagros Mts. (Lorestan and Chaharmahal and Bakhtiari provinces), along with those from the Shirkooh Mts., fell within sub-lineage IB. Last, multiple haplotypes from central Iran (Isfahan and Yazd Provinces), as well as Khuzestan, Kerman, and Hormozgan provinces, and a single specimen from the eastern slope of the Alborz Mts. (Semnan Province), were assigned to sub-lineage IIB.
Genetic diversity
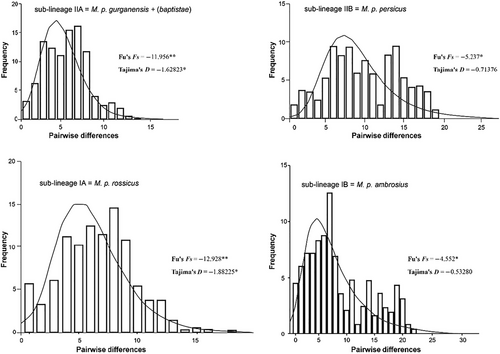
93 haplotypes were identifed among 145 cytb sequences included in this study, of which 42 were new within M. persicus populations. Haplotype and nucleotide diversity were the highest in sub-lineage IIB, followed by IB (Table 2). Clear geographic associations were evident for the haplotypes of sub-lineages IB and IIB. Within sub-lineage IB, haplotypes from the southern Zagros region (Shirkooh Mts.; Yazd province) and Bakhtiari region (Dopalan Mts.) are genetically divergent showing 14 and 10 mutations from all other specimens in central Zagros (Lorestan province), respectively, and within sub-lineage IIB, haplotypes from the southwestern Zagros region (Izeh; Khuzestan province) were separated by seven mutations from other counterparts. All four sub-lineages showed significant deviations from zero in Tajima's D test, the null hypothesis of constant population size. However, the Fu's Fs test displayed significance only for sub-lineages with larger sample sizes, (IA, n = 66; IIA, n = 30) (Table 2 and Fig. 3). According to the comparable Tau values, sub-lineages have experienced simultaneous allopatric diversification. The mean genetic divergence between lineage I and II was calculated at 10.3%, with divergence estimates between their respective sub-lineages being notably similar: 2.5% between sub-lineages IA and IB and 2.6% between IIA and IIB.
| IA | IB | IIA | IIB | |
|---|---|---|---|---|
| N | 66 | 23 | 30 | 26 |
| Nh | 31 | 17 | 21 | 24 |
| Np | 59 | 36 | 34 | 37 |
| Hd (SD) | 0.945 ± 0.014 | 0.972 ± 0.020 | 0.972 ± 0.015 | 0.982 ± 0.022 |
| Pi (SD) | 0.00641 ± 0.00 046 | 0.00958 ± 0.00 156 | 0.00542 ± 0.00 056 | 0.01020 ± 0.00 117 |
| K | 5.620 | 8.403 | 4.749 | 8.942 |
| Fu's Fs | −12.928** | −4.552* | −11.956** | −5.237* |
| Tajima's D | −1.88225** | −0.53280 | −1.62823* | −0.71376 |
| Tau | 3.835 | 3.548 | 3.492 | 5.516 |
| Theta 0–Theta 1 | 1.784–45.420 | 4.856–57.600 | 1.258–90.314 | 3.426–34.653 |
- “*” denotes the significant values in Fu's Fs and Tajima's D tests. N, sample size; Np, number of polymorphic sites; Nh, number of haplotypes; Hd, haplotype diversity; Pi, nucleotide diversity; K, the average number of nucleotide differences; Tau, scaled expansion time; Theta 0, population size before the expansion; Theta 1, population size after expansion.
Plague monitoring in Iran
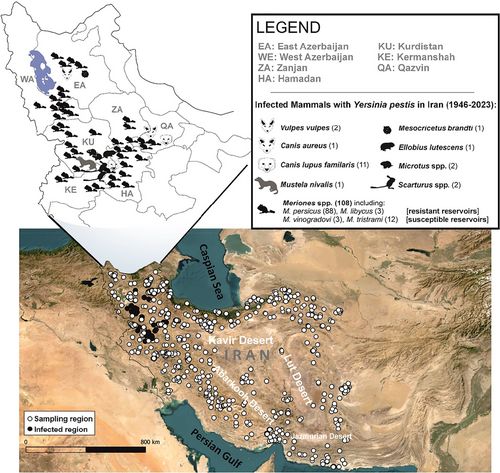
Over the past 77 years (1946–2023), nationwide animal plague surveillance in Iran has revealed plague infection in 127 mammals out of a surveyed animal population exceeding 17 000 (comprising small mammals and carnivores) (refer to Fig. 4). Among the surveyed mammals, rodents stand as the predominant group (n = 16647), with the genus Meriones being the most frequently captured (n = 10249). This establishes rodents as comprising 88.18% (n = 112) of the infected mammals, of which 94.64% (n = 106) pertain to the genus Meriones and 83.01% (n = 88) correspond to M. persicus.
DISCUSSION
Physio-geographic and climatic influences on the genetic diversity of M. persicus
The prominent physio-geographical features of the Iranian plateau, namely the Zagros and Alborz Mts., alongside the central Kavir and Lut deserts, have significantly impacted terrestrial vertebrate diversity (Yousefi et al. 2023), acting as either transition zones or physical barriers for various organisms (Yusefi 2021). The Zagros Mts., serving as a notable refuge during harsh climatic periods, have substantially influenced the biodiversity and genetic structure of regional fauna and flora (Rajaei et al. 2013; Dianat et al. 2017; Norozi et al. 2019; Kafash et al. 2020, 2021).
A significant negative deviation from the null hypothesis of constant population size indicated recent population growth across all M. persicus sub-lineages. According to the estimated divergence times, the final sub-structuring into recent sub-lineages predates the last glacial maximum, with sub-structuring occurring around 0.298 Mya between sub-lineages IA and IB, and 0.289 Mya between IIA and IIB (Dianat et al. 2017). Comparable Tau values among sub-lineages suggest simultaneous allopatric diversification across multiple refugia and postglacial expansions as credible explanations for the species’ current genetic composition and spatial distribution. The Kavir and Lut deserts separated sub-lineage IIA from IIB (Fig. 1b,c), reinforcing independent refugia in the northeast of the Alborz Mts. and the central massifs in Iran, respectively. Sub-lineage IIB's broad distribution from the central massifs to the Zagros Mts., reaching to the south of the Alborz Mts., implies that the Abarkooh desert has not served as a critical geographical barrier for this sub-lineage (Fig. 1b). North of the Zagros Mts. is proposed as a refugium for sub-lineage IA, while the Shirkooh Mts. in the south of the Zagros is suggested to have acted as a single refugium for sub-lineage IB (Dianat et al. 2017). However, considerable mutations separating haplotypes from the southern Zagros (Shirkooh Mts.; Yazd province) and the central Zagros Mts. (Lorestan and Bakhtiari regions) (Fig. 2b) imply that sub-lineage IB likely survived in at least two independent micro-refugia along the Zagros Mts.
Despite the scattered distribution of the four sub-lineages, the presence of sub-lineage IB deep within the range of sub-lineage IIB (see Fig. 1c) indicates a broader contact–parapatric zone, potentially due to recent population expansions (Fig. 3; Table 2) (Haddadian Shad et al. 2016; Dianat et al. 2017; Yusefi 2021; Yousefi et al. 2023). Moreover, the substantial genetic divergence (10.3%) between lineages I and II suggests the potential for their divergence into distinct species. This level of genetic divergence falls beyond what is typically found within intraspecific variation in rodents (Baker & Bradley 2006). On the whole, these variations may suggest a divergence at the species level. This proposition gains support from the cranial morphometric analysis illustrated in fig. 7 of Dianat et al. (2017) and an assessment of the basic field measurements presented in the current study (Table 1). Specifically, lineage I (potentially M. ambrosius) exhibits a smaller HBL, larger TL, and notably smaller HBL/TL when compared to lineage II (potentially M. persicus sensu stricto), even though could be partially due to sampling bias. Nevertheless, this hypothesis needs validation through additional evidence, particularly non-maternal nuclear markers or genome-wide approaches.
Subspecies taxonomy of M. persicus
Our findings largely corroborated the earlier results reported by Dianat et al. (2017). However, we made corrections to the affiliation of M. p. ambrosius, which was previously categorized under sub-lineage IIB. We also raised questions regarding the inclusion of M. p. baptistae, in sub-lineage IIA. Importantly, our study contributed significant clarity to the presumed distribution of each sub-lineage/subspecies in Iran (Fig. 1c), which will be outlined in detail subsequently.
Sub-lineage IIA is most likely taxonomically affiliated with M. p. gurganensis, given its inclusion of haplotypes from the type locality in Bojnourd, northeastern Iran. According to the current available data, we do not advocate the inclusion of M. p. baptistae in the same sub-lineage—unlike Dianat et al. (2017)—rather propose to sequence the materials from its designated type territory (Pasht Kuh; southwest Pakistan), a factor that could contribute significantly to the current uncertainty. Sub-lineage IIB, primarily occupying the Iranian central plateau, evidently links only to M. p. persicus, encompassing topotypes from the Qohrud Mts. This lineage spans from the south of the Alborz Mts. (Semnan province) southward to the central (Isfahan, Yazd, and Kerman provinces) and southern Zagros Mts. (Khuzestan, Hormozgan provinces). Sub-lineage IA is seemingly associated with M. p. rossicus (Armenia is the type locality) and found in high density, especially through the north of the Zagros and the western slope of the Alborz Mts. as well as in a few localities in the central Zagros Mts. (Lorestan and Isfahan provinces). Sub-lineage IB, formerly known solely from the Shirkooh Mts. in the central Iranian plateau—left unnamed by Dianat et al. (2017)—now appears to be a part of M. p. ambrosius, as closely clustered with the subspecies’ topotypes collected from Chaharmahal and Bakhtiari province (Thomas 1920). Sub-lineage IB has a more restricted range and is distributed exclusively through the central Zagros Mts. Thomas (1920) originally described this subspecies from central Zagros (Do Polan Mts. in Chaharmahal and Bakhtiari province), suggesting its range extended southward to the Persian Gulf coastline. Dianat et al. (2017) subsequently adopted this assumption, designating the southern populations from Hormozgan province as M. p. ambrosius (cf. fig. 2 in Dianat et al. 2017). Additionally, the absence of materials from the type locality led to the misclassification of M. p. ambrosius within the same sub-lineage with M. p. persicus (sub-lineage IIB in Fig. 3; Dianat et al. 2017). New data from the type territory of M. p. ambrosius reveals its distinctness from M. p. persicus and its sister relationship with M. p. rossicus. Additionally, M. p. ambrosius is also characterized by its relatively longer tail (Table 1), which is in line with its original description by Thomas (1920). Overall, new findings restrict the range of M. p. ambrosius from the central Zagros Mts. in the north to the Shirkooh Mts. in the south, excluding the Persian Gulf coastline. Further sampling is recommended from Fars and Ilam Provinces where genetic data are not available.
Enzootic plague and M. persicus subspecies
The persistence of plague within wildlife populations relies on reservoirs with high densities and metapopulation structures (Eisen & Gage 2009). A major plague reservoir in the Middle East is M. persicus, a common jird which exhibits relative resistance to infection (Baltazard et al. 1960) and notable intraspecific variation (Darvish 2011; Dianat et al. 2017; present study). Despite several epizootic outbreaks throughout the country (Shahraki et al. 2016), enzootic plague in Iran has been confined to the northern Zagros Mts. region, precisely aligning with the spatial distribution of M. p. rossicus (sub-lineage IA) (Fig. 1b). Meanwhile, Pasteur Institute's continuous surveillance in neighboring areas (Fig. 4)—such as the lower latitudes of the Zagros Mts. examining >4000 mammals—have yielded no evidence of infection in wildlife. As a result, enzootic plague in Iran is currently restricted to the northern Zagros, highlighting the possibility of a diverse role undertaken by different populations in plague epidemiology (Mahmoudi et al. 2021). Detecting enzootic cycles is challenging, especially during dormant phases that can span decades, during which the plague bacterium persists through localized microfoci and transitions between host populations (Stenseth et al. 2008; Barbieri et al. 2020; Mahmoudi et al. 2021). Thus, potential sampling biases within surveillance systems should not be overlooked. Thorough exploration along the Zagros Mts. is highly recommended due to intricate geo-climatic conditions and the coexistence of diverse sub-lineages. Our findings indicate that M. p. rossicus, which is found also in the Caucasus, eastern Anatolia, and Iraq (Denys et al. 2017), stands as the sole subspecies linked to the known enzootic outbreaks in Iran. Reports of plague infections in this species from the Caucasus region (Low pre-Araksinsk Mts. in Armenia and Azerbaijan) (Sludskyi 2014) suggest its potential to pose broader public health concerns within the region.
ACKNOWLEDGMENTS
Most of the samples of this study were taken during the regular monitoring of the plague research teams of the Pasteur Institute of Iran. In this regard, we appreciate the great help of Mr. Hamed Hanifi, Ali Mohammadi, and Seyyed Adel Hosseini during the field expedition.
CONFLICT OF INTEREST STATEMENT
We have no competing interests.




