Validation and development of eDNA metabarcoding primers for comprehensive assessment of Chinese amphibians
Dongyi Wu and Pingshin Lee are equal first authors.
Abstract
Environmental DNA (eDNA) metabarcoding has emerged as a powerful, non-invasive tool for biodiversity assessments. However, the accuracy and limitations of these assessment techniques are highly dependent on the choice of primer pairs being used. Although several primer sets have been used in eDNA metabarcoding studies of amphibians, there are few comparisons of their reliability and efficiency. Here, we employed lab- and field-tested sets of publicly available and de novo-designed primers in amplifying 83 species of amphibian from all three orders (Anura, Caudata, and Gymnophiona) and 13 families present in China to evaluate the versatility and specificity of these primers sets in amphibian eDNA metabarcoding studies. Three pairs of primers were highly effective, as they could successfully amplify all the major clades of Chinese amphibians in our study. A few non-amphibian taxa were also amplified by these primers, which implies that further optimization of amphibian-specific primers is still needed. The simultaneous use of three primer sets can completely cover all the species obtained by conventional survey methods and has even effectively distinguished quite a number of species (n = 20) in the Wenshan National Nature Reserve. No single primer set could individually detect all of the species from the studied region, indicating that multiple primers might be necessary for a comprehensive survey of Chinese amphibians. Besides, seasonal variations in amphibian species composition were also revealed by eDNA metabarcoding, which was consistent with traditional survey methods. These results indicate that eDNA metabarcoding has the potential to be a powerful tool for studying spatial and temporal community changes in amphibian species richness.
INTRODUCTION
Amphibians are the most threatened vertebrate group in the world, with more than 40% of known amphibian species currently categorized as being under some level of risk (Luedtke et al. 2023). Accurate information on amphibian geographical distributions and community composition is crucial for ecology and conservation biology. Traditional survey methods have played an important role in amphibian research, contributing greatly to the current understanding of global amphibian population dynamics and diversity patterns. However, substantial challenges persist, such as taxonomical challenges in distinguishing species (Padial et al. 2010) and inaccuracies in estimating the population density of amphibian species due to complexities in specific amphibian characteristics, life stage, and other factors (Guayasamin et al. 2015; Lopes et al. 2017). Considering current limitations posed by existing or traditional methods, approaches that could facilitate large-scale amphibian biodiversity assessment are urgently needed.
Environmental DNA (eDNA) and metabarcoding have emerged as powerful non-invasive tools for species detection in both temperate and tropical areas (Lopes et al. 2017; Sasso et al. 2017; Bálint et al. 2018; Li et al. 2021b; Sakata et al. 2022). Three primer sets are currently available for amphibian DNA metabarcoding, which use high-throughput sequencing to identify multiple taxa in complex samples (Taberlet et al. 2012; Zinger et al. 2019). For example, Batra_12S (batra_F and batra_R; Valentini et al. 2016; Table 1) could detect amphibian species from Europe (10 species, 7 genera, 4 families; Valentini et al. 2016), while BA_16S (BA-4445-F and BA-178-R; Table 1) could detect 25 southern American amphibian species (14 genera, seven families; Bálint et al. 2018). Mod/Vert_COI (Mod_RepCOI_F and VertCOI_7216_R; Reeves et al. 2018; Table 1) could detect three species of Hyla from the subtropical region of Florida. Another metabarcoding study of amphibians that used a different set of primers (Amph16) has demonstrated its effectiveness in detecting 16 amphibian species across a wide geographical region of Japan (Sakata et al. 2022).
| Custom name | Gene region | Primer names | Primer sequence 5′−3′ | Amplicon length (bp) | Annealing temperature | References |
|---|---|---|---|---|---|---|
| Batra_12S | 12s rRNA | batra_F batra_R |
For-ACACCGCCCGTCACCCT Rev-GTAYACTTACCATGTTACGACTT |
99 | 55°C | Valentini et al. (2016) |
| BA_16S | 16s rRNA | BA-4445-F BA-178-R |
For-RACCGTGCRAAGGTAGCR Rev-CCATRGGGTCYTCTCGTCT |
153 | 55°C | Balint et al. (2018) |
| Mod/Vert_COI | COI | Mod_RepCOI_F VertCOI_7216_R |
For-TNTTYTCMACYAACCACAAAGA Rev-CARAAGCTYATGTTRTTYATDCG |
244 | 48°C | Reeves et al. (2018) |
| Amph1_COI | COI | Chmf4 Ltest5 |
For-TYTCWACWAAYCAYAAAGAYATCGG Rev-AKAAYGGGYATGACYATRAAG |
199 | 48°C | This study, based on Che et al. (2012) |
| Amph2_COI | COI | Chmf4 Ltest2 |
For-TYTCWACWAAYCAYAAAGAYATCGG Rev-CRGGYTGRCTTARTTCWGC |
111 | 45°C | This study, based on Che et al. (2012) |
Whether the target taxa can be efficiently detected depends strongly on the sensitivity and versatility of the primers used. However, most metabarcoding primers are designed based on certain DNA regions of a limited number of taxa, which can potentially lead to amplification bias (Elbrecht & Leese 2015; Taberlet et al. 2018). For example, some primer detection efficiency may be limited to certain populations of vertebrate species but not to populations of the species inhabiting other regions due to high intraspecific genetic divergence (Elbrecht & Leese 2015; Zhang et al. 2020). Presently, no study has tested the performance of those primers to detect Gymnophiona in eDNA metabarcoding surveys, and only a few families of Anura and Caudata were tested whether they could be amplified by the DNA metabarcoding primers (Valentini et al. 2016; Sakata et al. 2022). Therefore, markers and primers should be validated for a range of target taxonomic groups before being applied in metabarcoding at a larger scale (Deagle et al. 2014). However, there are few comparisons of the reliability and efficiency of various sets of primers used. Only one recent study compared the 16S rRNA primers, but no comparison was made with primers that were developed based on other mtDNA regions (Sakata et al. 2022).
China harbors 650 species of amphibian belonging to three orders, 13 families, and 69 genera (AmphibiaChina 2023). A third of all Chinese anuran species are categorized as Near-threatened, Vulnerable, Endangered, or Critically Endangered (IUCN 2023). Accurate species identification in the field would greatly facilitate monitoring and conservation efforts (Wei et al. 2023). However, it is unclear whether previously published primer sets can amplify most of the amphibian families and detect amphibians in China. Therefore, the choice of which primers to use for rapid and effective detection of the local species of Chinese amphibians remains an open question.
Here, we tested the performance of sets of publicly available and de novo-designed primers in amplifying Chinese amphibians in both lab and field conditions, to evaluate the versatility and specificity of these primer sets in amphibian eDNA metabarcoding studies. A total of 96 individuals from all three Orders of Chinese amphibians (13 families, 45 genera, and 83 species) were used for primer testing. These species represented 100% of orders, 100% of families, and 65% of genera in China. First, we tested if the primers could successfully amplify all the major clades of Chinese amphibians. Second, we assessed the sensitivity and accuracy of the primer sets in detecting the residual DNA of Chinese amphibians in aquatic environments using samples collected from Wenshan National Nature Reserve. In this study, the best primers and experimental processes are proposed to provide a good theoretical basis for better application of eDNA in field surveillance.
MATERIALS AND METHODS
Primer selection and design
We selected three pairs of primers that are commonly used in amphibian metabarcoding studies (Table 1 and Fig. 1; Valentini et al. 2016; Bálint et al. 2018; Reeves et al. 2018). These primer sets amplified fragments within the 12S, 16S, and COI gene regions. To obtain universal primers specifically used for Chinese amphibian eDNA metabarcoding, we used the forward primers of Che et al. (2012) and designed two reverse primers based on this COI region. Primer design was performed using Primer Premier v.5.0 (Premier Biosoft International). The amplification lengths of these primer sets are 199 and 111 bp, respectively (Table 1 and Fig. 1).
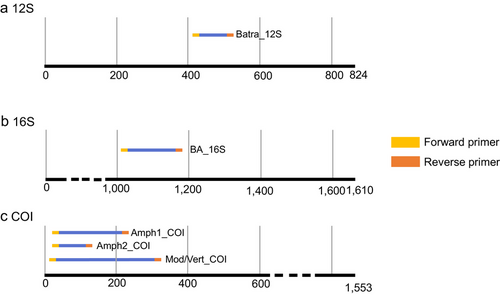
Evaluation of the versatility and specificity of primers
We assessed the versatility of the primer sets in amplifying the DNA from 96 individuals of amphibians (13 families, 45 genera, three orders: Anura, Caudata, and Gymnophiona). Two invertebrates and five vertebrates that represented a diverse range of invertebrates and vertebrate classes were tested for primer specificity (Table S1, Supporting Information). All samples were collected from non-invasive cotton swabs or roadkill samples and were subsequently stored in 75% and 95% anhydrous ethanol solutions, respectively. DNA was extracted from the samples using the DNeasy Blood and Tissue Kit (QIAGEN Science, Hilden, Germany) following the manufacturer's protocol before storage at −20°C. Subsequently, the extracted DNA from each sample was diluted to a concentration of 1 ng μL−1, and further dilutions were made 100, 1000, and 10 000 times, respectively (i.e. 1, 1 × 10−2, 1 × 10−3, 1 × 10−4 ng μL−1).
Two newly designed COI primer sets (Amph1_COI and Amph2_COI) were used in gradient PCR at Ta in the range of 42–60°C (the amplification procedure is as below) according to Che et al. (2012). PCR products were visualized on a 1.5% agarose gel to determine the optimal Ta for each primer set (Table 1).
PCR amplification was performed using the five primer sets mentioned above. PCR conditions were as follows: initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 45 s, annealing under a temperature range of (Ta) for 45 s (Table 1), and extension at 72°C for 30 s; and final extension at 72°C for 10 min, followed by storage at 4°C. All PCR reactions were repeated three times. The final PCR products were visualized on a 1.5% agarose gel under UV light. In addition, for COI primer sets that showed poor amplification effect, nested-PCR optimization was performed for all the samples. In the first round of PCR, a pair of primers that amplified a longer fragment was used, and the amplicon was used as a template for another primer pair that amplified a shorter fragment in the second round of PCR. Here, we performed two nested-PCR using a combination of three COI primer sets: Amph1_COI + Amph2_COI and Mod/Vert_COI + Amph2_COI (Table S1, Supporting Information). Four negative controls were included in each PCR run.
Field survey and construction of eDNA reference database
Wenshan National Nature Reserve is one of the reserves with the richest biodiversity and the highest number of rare plant and animal species. The reserve is known for its unique ecological features and is home to a variety of amphibians (Yang et al. 2008). In this study, nine sampling sites within Wenshan National Nature Reserve were surveyed twice. The first survey was conducted in November 2021 and the second survey was conducted in February 2022. The sites were mainly set at the intersection of water and habitats suitable for amphibians. We performed the traditional line transect method and the quadrat method during the field investigation of sampling sites (Fig. S1 and Table S2, Supporting Information). From 2000 to 2400, three investigators surveyed the sampling sites at an average speed of 1 km h−1. The species and number of amphibians were recorded at each site through visual observation, and acoustics were performed to supplement the species detected along the transect line. To avoid repetition of recording the same individuals through both sampling methods, only individuals who were observed but not heard, or individuals who were not observed but had callings heard in the same zone were recognized as distinct individuals. Non-invasive swabs were obtained from observed amphibian larvae and tadpoles that had not yet fully metamorphosed, and the swabs were stored in 95% alcohol. DNA was extracted from the collected swabs using DNeasy Blood and Tissue Kit (QIAGEN Science, Hilden, Germany) and amplified by PCR using the COI primers (Chmf4 and Chmr4) (Che et al. 2012). We sent the PCR products to Beijing Tsingke Biotech Co., Ltd. for Sanger sequencing. The sequences were blasted against the COI database of the AmphibiaChina web (AmphibiaChina 2023) to accurately identify amphibian species based on 97% sequence similarity.
To ensure the accuracy of taxonomic assignment to sequences recovered from environmental samples based on 29 amphibians recorded in the reserve (Yang et al. 2008), we constructed a reference database of local amphibians. This database was built using samples collected from field surveys and preserved amphibians in the laboratory (Table S3, Supporting Information). Here, we constructed three databases based on PCR amplification of 16S rRNA, 12S rRNA, and COI mitochondrial genes from each sample using BA_16S, Batra_12S, and COI primers (developed by Che et al. 2012), respectively. All PCR products were then sent for Sanger sequencing.
eDNA field test
In November 2021 and February 2022, water samples were collected from nine sampling sites between 9:00 and 15:00 (Fig. S1 and Table S2, Supporting Information). For the line transect survey method, both sampling times commenced at the initial point of each line transect. For the quadrat survey method, both times of water sample collection were performed at a fixed position of each quadrat. At each sampling point, 2 L of surface water was collected using a plastic sampler that had been previously sterilized with 10% sodium hypochlorite. To ensure the reproducibility of the research results, two replicate water samples were collected at each collection point. All collected water samples were filtered directly using 0.45-μm mixed cellulose ester filters (MCE; Whatman International Ltd.) at each sampling site to ensure the rapid enrichment of water DNA on the filter membrane. The filtration membranes were then stored in 95% anhydrous ethanol and brought back to the laboratory for DNA extraction. To avoid cross-contamination, all water collection and filtration equipment has been sterilized with 10% sodium hypochlorite.
DNA was extracted using DNeasy Blood and Tissue Kit (QIAGEN Science, Hilden, Germany) and stored at −20°C. To check for possible exogenous DNA contamination during the experiment, ddH2O was used as a negative control for DNA extraction and PCR. Three candidate primer sets (Batra_12S, BA_16S, and Amph1_COI + Amph2_COI) could successfully amplify the target gene fragments of 96 amphibians at a concentration of 1 ng μL−1 (see Result). Therefore, we employed these primer sets to amplify the mitochondrial fragments of the eDNA samples. The PCR amplification conditions and process were as above, with three replicates for each sample. PCR products of the same primer set in each group were collected in equal volumes, and the target DNA was visualized on a 1.5% agarose gel. All DNA extraction and amplification took place in separate lab rooms. The library was sequenced (2 × 150 bp PE) using the Illumina NovaSeq 6000 system (Illumina Inc.), and the expected total number of reads for each sample was set to 500 000. Library preparation and paired-end sequencing were conducted by Shanghai Biozeron Biotechnology CO., LTD.
Bioinformatics analysis
In this study, all obtained Sanger sequence data were qualitatively detected and manually proofread by Seq-Man in LASERGENE 7.0. Sequences were then aligned using MAFFT v.7.110 (Katoh & Standley 2013) with default settings (E-INS-i algorithm).
We checked the quality of high-throughput sequence data using Fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmomatic v.0.36 (Bolger et al. 2014) programs were used to trim paired-end reads with a sliding window of 10 bp. When the average quality value in the window was <20, the following bases were cut off and the sequences with Phred quality scores less than 20 were filtered. Flash v.1.2.11 (Magoč & Salzberg 2011) was used to merge paired-end reads with an overlap length of 10 bp, and the maximum allowed mismatch ratio in the overlap area is 0.2. Sequences were clustered into operational taxonomic units (OTUs) at 97% identity using USEARCH v.10 (Edgar 2010). Any chimeric sequence was removed in the clustering process. The sequence with the highest abundance in each OTU was selected as the representative sequence. We used the Blastn tool (Camacho et al. 2009) and the reference database customized for this study to classify OTUs with ≥97% identity and ≤10−5 E-value into taxonomic groups. MEGA v.6.0.6 (Tamura et al. 2013) was used to construct Neighbor-joining trees (NJ) for OTUs and reference species sequences from the database. Mean genetic distances between and within species were calculated based on uncorrected Kimura 2-parameter in MEGA v.6.0.6 (Tamura et al. 2013).
Statistical analysis of eDNA data
We categorized the detected amphibian species based on habitat and ecological types using species descriptions from the IUCN (2023). The categories include Semi-aquatic, Terrestrial, and Arboreal types. Venn diagrams and heat maps were created to compare the number of species detected by different primer sets as well as eDNA metabarcoding and traditional surveys. To evaluate the feasibility of different methods for amphibian field investigations, we compared the species detection rates from different eDNA primer sets and traditional surveys. To evaluate whether eDNA data from different primer sets could reflect the relative abundance of amphibians in the wild, we plotted a combination map based on eDNA reads and species abundance from traditional surveys. We used ordinary least squares (OLS) regression and Pearson correlation coefficient analysis to assess the correlation between eDNA reads and species abundance. To optimize the model fit, we performed a logarithmic transformation (log10) on the eDNA data. The linear relationship between eDNA reads and species abundance was fitted using the lm function. All statistical analyses were performed using R v.4.1.3 (R Core Team 2022).
RESULT
Primer specificity and versatility
We tested the specificity of five primer sets in detecting amphibians using seven non-amphibian taxa (two invertebrates and five vertebrates; Table 1). The results showed that all four groups of primers amplified non-amphibian taxa, but the amplification effects of these primer sets on different DNA concentrations varied (Fig. 2a). Batra_12S, BA_16S, Amph2_COI, Amph1_COI + Amph2_COI, and Mod/Vert_COI + Amph2_COI could detect all the species above at all tested template concentrations of 1, 1 × 10−2, and 1 × 10−3 ng μL−1. Meanwhile, BA_16S, Amph1_COI + Amph2_COI, and Mod/Vert_COI + Amph2_COI could amplify the DNA of certain species at a concentration of 1 × 10−4 ng μL−1. Amph1_COI had the weakest amplification sensitivity, which could only amplify the DNA template of the target species at 1 ng μL−1 (Fig. 2a).
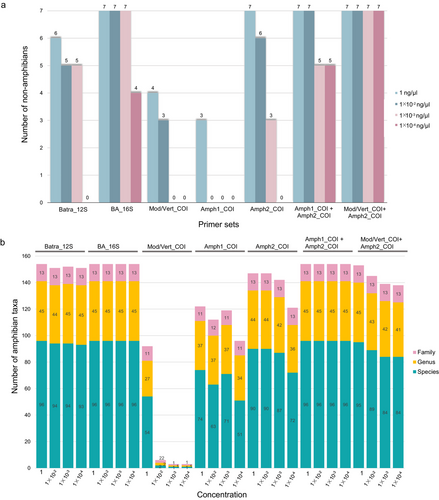
When comparing the amplification success of different primer sets for Chinese amphibians, Batra_12S, BA_16S, and Amph1_COI + Amph2_COI could all amplify 100% of the target fragments of 96 species at a DNA concentration of 1 ng uL−1, while Mod/Vert_COI could only amplify 56% of the individuals (Fig. 2b; Table S1, Supporting Information). The BA_16S and Amph1_COI + Amph2_COI could both amplify 100% of all species when the template DNA concentration was diluted to a low multiple, while the amplification efficiency of Batra_12S reached more than 95% for different concentrations. Mod/Vert_COI showed the lowest amplification success rate at the four concentrations (56%, 2%, 1%, and 1%, respectively) (Fig. 2b; Table S1, Supporting Information).
Field amphibian survey and reference database
We found 10 species of amphibians in the two traditional surveys in total, which belonged to five families and 10 genera (Table 2). In November 2021, seven Anuran and one Caudata species were recorded. Bufo gargarizans (n = 23) and Nidirana pleuraden (n = 20) had the highest number of individuals detected. Among the six Anuran species surveyed in February 2022, Quasipaa boulengeri was recorded the most often (n = 14). Ten amphibian samples were collected from two traditional surveys, and 19 samples were preserved in the laboratory. We obtained a total of 29 known amphibians from Wenshan National Nature Reserve to construct three reference databases (Yang et al. 2008). Three reference databases consisting of three different genetic markers (12S, 16S, and COI) were constructed from 29 amphibians, and the length of each gene was 100, 153, and 686 bp, respectively. Sequence fragments amplified by different eDNA universal primer sets were uploaded to their respective databases (Table S3, Supporting Information).
| Family | Genus | Species | Habitat and ecology | Batra_12S | BA_16S | Amph1_COI + Amph2_COI | Traditional monitoring |
|---|---|---|---|---|---|---|---|
| Salamandridae | Tylototriton | Tylototriton yangi | Semi-aquatic | + | + | − | + |
| Megophryidae | Leptobrachella | Leptobrachella feii | Semi-aquatic | + | + | − | − |
| Megophryidae | Leptobrachella | Leptobrachella bourreti | Semi-aquatic | + | + | + | + |
| Megophryidae | Leptobrachium | Leptobrachium ailaonicum | Terrestrial | + | + | + | + |
| Megophryidae | Boulenophrys | Boulenophrys daweimontis | Semi-aquatic | − | + | − | − |
| Megophryidae | Boulenophrys | Boulenophrys jingdongensis | Semi-aquatic | + | + | − | + |
| Megophryidae | Xenophrys | Xenophrys maosonensis | Semi-aquatic | − | + | + | − |
| Bufonidae | Bufo | Bufo gargarizans | Terrestrial | + | + | + | + |
| Bufonidae | Duttaphrynus | Duttaphrynus melanostictus | Terrestrial | + | + | − | − |
| Ranidae | Nidirana | Nidirana pleuraden | Semi-aquatic | + | + | + | + |
| Ranidae | Odorrana | Odorrana grahami | Semi-aquatic | + | + | + | + |
| Ranidae | Rana | Rana chaochiaoensis | Terrestrial | + | + | + | + |
| Rhacophoridae | Zhangixalus | Zhangixalus puerensis | Arboreal | + | + | + | − |
| Rhacophoridae | Kurixalus | Kurixalus lenquanensis | Arboreal | + | − | − | − |
| Rhacophoridae | Feihyla | Feihyla palpebralis | Arboreal | − | − | + | − |
| Dicroglossidae | Fejervarya | Fejervarya multistriata | Semi-aquatic | + | + | − | − |
| Dicroglossidae | Quasipaa | Quasipaa boulengeri | Semi-aquatic | + | + | + | + |
| Dicroglossidae | Nanorana | Nanorana yunnanensis | Semi-aquatic | + | + | + | + |
| Microhylidae | Microhyla | Microhyla pulchra | Terrestrial | − | − | + | − |
| Microhylidae | Microhyla | Microhyla fissipes | Terrestrial | + | − | − | − |
Sequencing result of eDNA field samples
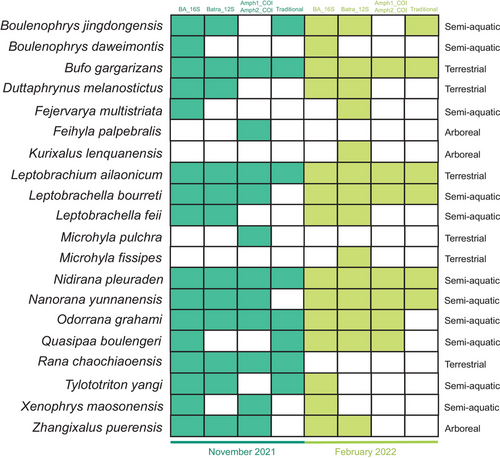
A total of 36 water samples were collected from the nine sampling points during two different seasons. Agarose gel electrophoresis results showed no PCR amplification for all negative controls, indicating that there was no contamination during the experiment. After the screening process, for the first batch of samples that were collected, a total of 3 487 163 effective sequence reads were obtained based on the amplification of Batra_12S, with an average length of 72 bp; BA_16S obtained 5 530 827 effective reads with an average length of 114 bp; Amph1_COI + Amph2_COI obtained 3 367 395 reads. For the second collection, based on the amplification of Batra_12S, BA_16S, and Amph1_COI + Amph2_COI, 3 097 788, 5 362 803, and 1 049 170 reads were obtained, respectively. The average length of sequence reads generated from each primer set was consistent with the result of the first collection. After subsequent processing, 511, 582, and 723 OTUs of Batra_12S, BA_16S, and Amph1_COI + Amph2_COI were identified as amphibians from the sampling event in November 2021. Following further classification and identification of the OTUs, 18 amphibian species belonging to seven families and 16 genera were discovered from eDNA metabarcoding of the three primer sets. For the samples collected in February 2022, 517, 713, and 1296 OTUs of the three primer sets were identified as 17 amphibian species, belonging to seven families and 15 genera (Fig. 3 and Table 2).
The constructed NJ trees showed that the OTUs clustered together with candidate species from the reference database with strong support (Figs S2–S4, Supporting Information). The genetic distance between all tested OTUs and reference species was smaller than the genetic distance between two closely related species.
Effectiveness of eDNA metabarcoding primer sets in field investigation
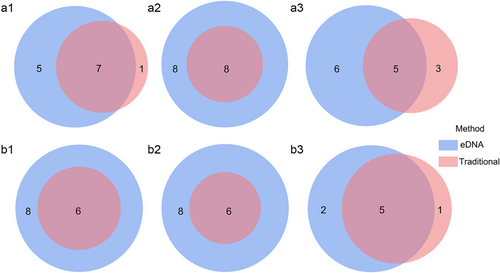
A total of 20 amphibian species, belonging to seven families and 17 genera, were detected using eDNA and traditional surveys (Table 2). Both Batra_12S and BA_16S detected 16 amphibians belonging to 15 genera and six families; 14 species were detected by the two primer sets. The non-overlapping species detected by Batra_12S and BA_16S were Kurixalus lenquanensis and Microhyla fissipes, Boulenophrys daweimontis, and Xenophrys maosonensis, respectively. Amph1_COI + Amph2_COI detected 12 amphibian species belonging to 12 genera and 6 families (Table 2). During the two sampling periods, the composition of detected species varied among different primer sets, but all species with Arboreal, Terrestrial, and Semi-Aquatic habitat types were detected (Fig. 3). Overlapping species between primer sets were not counted; however, BA_16S independently detected B. daweimontis and Fejervarya multistriata, while Amph1_COI + Amph2_COI independently detected Microhyla pulchra during the first sampling period (Fig. 3). For the sampling in February 2022, three unique species—F. multistriata, M. fissipes, and K. lenquanensis were detected with Batra_12S primers (Fig. 3), while B. daweimontis, X. maosonensis, and Tylototriton yangi were exclusively identified with BA_16S primers (Fig. 3). The arboreal species were detected only by the Batra_12S and BA_16S primer sets (Fig. 3).
When comparing the effectiveness of eDNA metabarcoding and the traditional method in amphibian monitoring, it was observed that all levels of species richness detected with different eDNA metabarcoding primer sets were higher than those detected by the traditional survey method (Fig. 4a,b). Among them, BA_16S primers covered all the species detected from the traditional survey. Batra_12S also identified all amphibian species recorded from the traditional survey, excluding Q. boulengeri. The amphibians detected by Amph1_COI + Amph2_COI did not include the three species identified by the traditional survey method—T. yangi, Boulenophrys jingdongensis, and Q. boulengeri (Fig. 4a,b and Table 2).
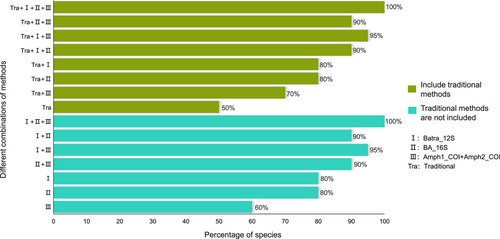
The results of comparing species detection rates of both methods showed that the eDNA metabarcoding combined with the use of three different genetic markers could detect 100% amphibians in the survey area, regardless of the traditional survey method (Fig. 5). The species coverage rate of the eDNA method with two combinations of different genetic markers was consistent with the total species coverage rate of both eDNA metabarcoding and traditional surveys (Fig. 5). In addition, the species coverage rate with any of the three primer sets was higher than that of the traditional methods; even Amph1_COI + Amph2_COI (60%) with the lowest detection rate detected a higher number of species than traditional surveys (50%) (Fig. 5).
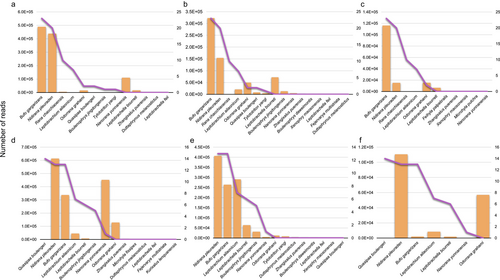
All three primer sets could accurately detect amphibians in the field; the amphibians detected by different primer sets had different sequence abundance and read count (Fig. S5, Supporting Information). Figure 6 shows the comparison of the abundance of amphibian species detected by each primer set and the traditional survey. B. gargarizans and N. pleuraden detected by the eDNA method showed relatively high sequence abundance, as compared to the species abundance recorded by the traditional method, and they were the dominant species in the survey area. The sequence abundance of some species (e.g. Leptobrachella bourreti, Odorrana grahami, and Nanorana yunnanensis) detected by eDNA could not fully reflect the species abundance recorded from traditional survey methods. OLS and Pearson correlation coefficient analysis showed that the numbers of reads from certain primer sets are positively correlated with individuals of a species. However, such correlations are susceptible to random errors (Batra_12S: P = 0.28, r2 = 0.02; Amph1_COI + Amph2_COI: P = 0.94, r2 = 0.001; Fig. S6, Supporting Information). Therefore, it is best practice to involve more replications and the use of multiple markers in future detection.
DISCUSSION
Efficiency and specificity of eDNA metabarcoding primers tested in lab
The choice of metabarcoding primers is one of the most critical factors in determining the detection probability of specific taxonomic groups for eDNA studies (Freeland 2017; Alberdi et al. 2018). The ideal primers must be universal enough to amplify a large proportion of the target taxonomic group, and their efficiency should be tested before field application. In this study, a new pair of modified COI primer sets (Chmf4 + Ltest5 and Chmf4 + Ltest2), in combination with the previously published 16S (BA-4445-F + BA-178-R) and 12S primers (batra_F + batra_R), effectively amplified target DNA from 96 individuals of Chinese amphibians belonging to 45 genera and 13 families at extremely low quality (1, 1 × 10−2, 1 × 10−3, and 1 × 10−4 ng μL−1). This suggests that all of these primers are suitable candidates as eDNA metabarcoding primers for large-scale amphibian surveys in China. Additionally, China and its neighboring countries such as those in eastern Asia (Japan, Korea) and Indochina (Vietnam, Laos, Myanmar, and Thailand), which share similar amphibian families, could use these eDNA metabarcoding primers for biodiversity surveys. For example, all the 13 families tested in this study represented all the families of amphibians from Japan, Myanmar, Thailand, Laos, and Vietnam, and 93% of families from Korea (Table S4, Supporting Information).
Furthermore, ideal eDNA metabarcoding primers theoretically should have taxonomic specificity for a diverse range of targeted taxa while showing minimal or no co-amplification of non-target groups (Alberdi et al. 2018; Collins et al. 2019). Our research has revealed that the primers for all three genes have low specificity for amphibians, as their detection includes other animal groups (birds, mammals, fish, and insects; Table S1, Supporting Information). This result is generally consistent with other metabarcoding studies (e.g. fish, Zhang et al. 2020), where no primers could successfully eliminate non-specific amplification for other groups. The elimination of non-specific amplification will require researchers to perform library analysis of non-target species, which often results in higher costs of eDNA studies. This suggests that further optimization of amphibian-specific primers is still needed in future applications. However, despite their lower specificity, the primers tested in this study can still achieve amplification of most targeted amphibian groups. Moreover, due to their non-invasiveness and high sensitivity, they have great potential as tools for the rapid assessment of species diversity. These non-specific results can also provide more valuable information for species diversity assessment of ecological communities to some extent.
Field evaluation of eDNA universal primers and implications for amphibian monitoring
Our eDNA metabarcoding study showed that universal primers have great potential to reveal the species richness of amphibians in the field. The non-invasive eDNA survey with the use of three pairs of primers can completely cover the species obtained by conventional survey methods and effectively distinguish the species in the study area (Fig. 4). The species detected by eDNA not only completely covered the species identified by conventional surveys in nine sampling sites (see Fig. S7, Supporting Information) but also encompassed all the species in two different seasons (Fig. 4). It is worth noting that the number of species detected by the eDNA method is higher than that by the conventional survey method. Furthermore, our research has demonstrated that, despite variability in habitat selection among amphibians, the eDNA method remains effective in detecting species with less contact with water, such as terrestrial and arboreal species. This may be due to the eDNA methods being sensitive enough to detect rare species even at their elusive life stages, such as tadpoles. Additionally, it can be detected during the non-breeding season when amphibians are difficult to observe (Deiner & Altermatt 2014; Sasso et al. 2017; Sakata et al. 2022). This probably explains why M. fissipes, B. daweimontis, X. maosonensis, and so on failed to be detected by traditional methods in this study (Table 2). Besides, seasonal variations in species composition of the nature reserve were also revealed by eDNA metabarcoding, and they showed consistencies in results with that of traditional survey methods. These results indicate that eDNA metabarcoding has the potential to be a powerful tool for studying the spatial and temporal community changes in amphibian species richness.
In this study, no single primer set alone was able to cover all species of a region, and multiple primers are required for surveying Chinese amphibians. The 16S and 12S primers were able to detect 80% of the species from the nine sampling sites (Fig. 5). The COI primers were less effective and could only detect 60% of the species. Additionally, the results showed that each pair of primers amplified a unique set of species in each season (Fig. 4a,b). For example, Feihyla palpebralis and M. pulchra were detected only by COI primers in November, but not by other primers. The difference in species composition detected by each primer set may be due to bias in primer detection toward different species in different environments (Belle et al. 2019; Weigand et al. 2019). Previous amphibian diversity assessments using eDNA metabarcoding have mainly used single universal primers, which may lead to an underestimation of the local amphibian diversity (Valentini et al. 2016; Lopes et al. 2017; Li et al. 2021b; Sakata et al. 2022). We suggested it would be best to use all three-pair primer sets in detecting all species of a region. However, if the funding is limited, primer Batra_12S or primer BA_16S could be used as a preliminary amphibian survey that could detect at least 80% of the species in a region, including most of the terrestrial and arboreal species.
Previous studies have discussed the high controversy on whether COI is suitable for eDNA metabarcoding (Collins et al. 2019; Zhang et al. 2020). Due to the variability in the COI region, finding conserved priming regions among short target fragments (<300 bp) is difficult. This raised concerns about the suitability of some COI primers considering their high species-specific primer–template mismatches, which can result in inefficient, biased amplifications that may hinder downstream analyses (Deagle et al. 2014; Elbrecht & Leese 2015). However, our results here confirmed that COI primers can effectively amplify all amphibian species in China and detect certain specific taxa of species in water environments (Fig. 3; Table 2). Moreover, the COI gene has the highest species coverage in the database (BOLD, Barcode of Life Database; Ratnasingham & Hebert 2007), and this makes COI a promising standard marker for eDNA metabarcoding of amphibians.
In addition, our results show that the detection of all three primer pairs cannot fully reflect the seasonal changes in the abundance of most amphibian species (Fig. 6). The relative sequence abundance detected by different makers and conventional survey methods varied for certain species, such as N. yunnanensis and O. grahami (Fig. 6). Only a few dominant species, such as B. gargarizans and N. pleuraden, had detected a read abundance of eDNA that corresponded to the abundance of species surveyed by the traditional method. Due to the complexity of eDNA ecology in natural water environments (Barnes & Turner 2016), accurately estimating the relative biomass of species based on eDNA read abundance is challenging (Ushio et al. 2018). Although some previous studies of eDNA metabarcoding showed a positive relationship between species abundance and sequence read abundance (Klobucar et al. 2017; Ushio et al. 2018; Li et al. 2021a), many other studies produced inconsistent results (Maruyama et al. 2014; Evans et al. 2015; Shu et al. 2021). Resolving problems such as PCR bias, sequencing errors, and bioinformatics analysis system errors will contribute to eDNA investigations of species abundance in the future (Evans & Lamberti 2018; Lacoursière-Roussel & Deiner 2021).
ACKNOWLEDGMENTS
We are grateful to Dr. Marcio Roberto Pie for providing language refinements and offering valuable insights on the article. We thank Xiaolong Liu, Renda Ai, Zezhao Kang, Rui Chen, Yang Zhang, and Jiangyu Li for their help in the eDNA sample collection and during the experiments. This work was supported by grants from the National Natural Science Foundation of China (NSFC32170478, NSFC32001222, NSFC32060828, NSFC32370478) to Z.Y. and P.L., the Fundamental Research Funds for the Central Universities (SWU-KR22014), the “Special Fund for Youth Team of Southwest University” (SWU-XJPY202302), “Youth Top Talent Program of Chongqing” (CQYC 20220510893) and Conservation and Genetic Germplasm Creation of Aquatic Resources in the Upper Yangtze River (2010423005) to Z.Y.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
Open Research
DATA AVAILABILITY STATEMENT
All data generated in this study have been reported within the main text or in the supporting information.




