Comparative genomics provides insights into molecular adaptation to hypermetamorphosis and cantharidin metabolism in blister beetles (Coleoptera: Meloidae)
Abstract
Blister beetles (Coleoptera: Meloidae) are currently subdivided into three subfamilies: Eleticinae (a basal group), Nemognathinae, and Meloinae. These are all characterized by the endogenous production of the defensive terpene cantharidin (CA), whereas the two most derived subfamilies show a hypermetamorphic larval development. Here, we provide novel draft genome assemblies of five species sampled across the three blister beetle subfamilies (Iselma pallidipennis, Stenodera caucasica, Zonitis immaculata, Lydus trimaculatus, and Mylabris variabilis) and performed a comparative analysis with other available Meloidae genomes and the closely-related canthariphilous species (Pyrochroa serraticornis) to disclose adaptations at a molecular level. Our results highlighted the expansion and selection of genes potentially responsible for CA production and metabolism, as well as its mobilization and vesicular compartmentalization. Furthermore, we observed adaptive selection patterns and gain of genes devoted to epigenetic regulation, development, and morphogenesis, possibly related to hypermetamorphosis. We hypothesize that most genetic adaptations occurred to support both CA biosynthesis and hypermetamorphosis, two crucial aspects of Meloidae biology that likely contributed to their evolutionary success.
INTRODUCTION
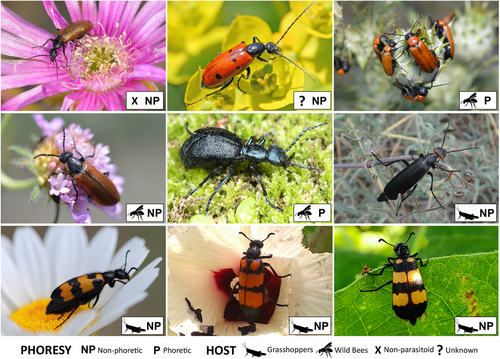
Meloidae, commonly known as blister beetles, is a cosmopolitan family of beetles with approximately 3000 species and 130 genera. They are found globally, except in New Zealand, certain Polynesian Islands, and Antarctica, and are classified into three subfamilies: Eleticinae, Nemognathinae, and Meloinae (Bologna et al. 2010; Riccieri et al. 2022, 2023). Blister beetles are all characterized by the endogenous production of a toxic terpene called cantharidin (CA), which is exuded through reflex-bleeding as a defensive strategy against predators and used to protect the eggs after oviposition (Carrel & Eisner 1974). Notably, males of canthariphilous species related to Meloidae, such as Anthicidae and Pyrochroidae, sequester CA for use as an attractant during courtship (Eisner et al. 1996; Molfini et al. 2023). All Meloidae, with the possible exception of Eleticinae (Pinto et al. 1996; Bologna et al. 2001; Bologna & Di Giulio 2011), typically undergo hypermetamorphic larval development, featuring four morphologically distinct larval stages, each serving different functions related to host-finding, dispersal, and body growth (Bologna et al. 2010; Bologna & Di Giulio 2011). Blister beetles also show a wide range of lineage-specific ecological adaptations, especially in their larval habits, such as the way of attaining the food sources (i.e. by phoresy or crawling) and the preferred host (i.e. wild bees or grasshoppers) (Bologna et al. 2010; Bologna & Di Giulio 2011).
So far, most of the genomic sequencing efforts have been directed toward species of blister beetles belonging to the Meloinae subfamily, for example, Hycleus phaleratus (Pallas, 1781), Hycleus cichorii (Linnaeus, 1758) (Wu et al. 2018; Zhou et al. 2023), Epicauta sibirica (Pallas, 1773) (reported as chinensis Laporte, 1849, but see Liu et al. 2016; Tian et al. 2021) to provide reference data and inspect genes involved in CA de novo biosynthesis.
Through this work, we aimed to highlight the molecular basis of the shared features of Meloidae and unravel the genetic novelties underlying the wide array of lineage-specific adaptations. To this aim, we conducted a genomic comparative analysis through an extensive sampling within Meloidae. To do so, we newly generated genomes of five species of blister beetles showing diverse larval ecological adaptations concerning phoresy and host selection, sampled across the three extant Meloidae subfamilies (Fig. 1). Other resources from already published blister beetle genomes were also considered, as well as those from Pyrochroa serraticornis (Scopoli, 1763), a closely-related canthariphilous species (Molfini et al. 2023), which was included for a more comprehensive investigation on the common genomic features related to CA metabolism.
MATERIALS AND METHODS
Sample collection
Five specimens (each belonging to a different species with a peculiar ecology)—collected on field between 2005 and 2021, preserved in 96% ethanol and stored at 4°C in the collection of M.A. Bologna (MAB) at Roma Tre University—were identified using an Olympus SZX12 Stereomicroscope and selected for this work (Fig. 1): (1) Iselma pallidipennis Haag-Rutenberg, 1879 (Eleticinae, Derideini; collected from South Africa, Northern Cape, 31°22′19.8″S, 19°01′50.8″E), non-phoretic, non-parasitoid (Bologna et al. 2001); (2) Stenodera caucasica (Pallas, 1781) (Nemognathinae, Stenoderini; collected from Turkey, Sinop Province, 41°39′42.3″N, 34°52′29.1″E), non-phoretic, with unknown hosts (Bologna et al. 2002); (3) Zonitis immaculata (A.G. Olivier, 1789) (Nemognathinae, Nemognathini; collected from Italy, Calabria, 39°20′31.4″N, 16°13′08.6″E), phoretic and parasitoid of wild bees (Bologna et al. 2010); (4) Lydus trimaculatus (Meloinae, Lyttini; collected from Italy, Tuscany, 42°51′25.0″N, 11°53′46.5″E), non-phoretic, parasitoid of wild bees (Bologna et al. 2010); (5) Mylabris variabilis (Meloinae, Mylabrini; collected from Italy, Sardinia, 40°34′20.9″N, 9°38′22.7″E), non-phoretic, parasitoid of grasshoppers (Pan & Bologna 2014).
Genomic sequencing, assembly, and annotation
Genomic DNA from the whole body was extracted using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions. Genomic DNA integrity was assessed using agarose gel electrophoresis (1%). The concentration and purity of the extracted DNA samples were evaluated using Nanodrop (Thermo Scientific).
For each species, whole-genome libraries were constructed and paired-end sequenced (2 × 150 bp) on an Illumina NovaSeq 6000 platform (Eurofins Genomics Europe, Konstanz, Germany).
Raw sequences were processed using Trimmomatic v0.39 (Bolger et al. 2014) to remove adapters and filter out low-quality bases. Reads shorter than 50 nucleotides were discarded. Potentially contaminated reads were removed using Kraken2 v2.1.2 (Wood et al. 2019) using a custom database including the following taxa: archaea, bacteria, fungi, human, protozoa, and viruses.
The program Platanus v1.2.4 (Kajitani et al. 2014) was used to de novo assemble each species’ genome by applying three main steps: (a) contigs were assembled using paired-end clean reads (67-mer extension procedure); (b) contigs were joined together in a scaffold using paired-end information; (c) scaffolds were processed using a gap-closing step to minimize unknown bases. Contigs/scaffolds smaller than 500 bp were discarded. The completeness of each genome assembly was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) v5.5.0 (Insecta_odb10 database; 1367 genes). The newly assembled genomes were deposited in GenBank (JAZBGW000000000, JAZBGX000000000, JAZBGY000000000, JAZBGZ000000000, JAZBHA000000000).
The clean reads were used to estimate the genome size of the five species based on k-mer frequency (k-mer value: 21). To this end, the program Jellyfish v2.2.6 (Marçais & Kingsford 2011) was used to count the occurrence of each k-mer in the sequence set of each species. The Jellyfish output was then processed by GenomeScope v2.0 (Ranallo-Benavidez et al. 2020) to estimate genome heterozygosity, repeat content, and genome size.
To functionally annotate protein-coding genes in the genome assembly of the five species, we used the Funannotate v1.8.4 pipeline (https://zenodo.org/records/4054262) in a conda computing environment. Briefly, Funannotate provides a modular workflow to generate a soft-masked genome and predict gene models using different sources of evidence that, in a final step, are merged by the program EvidenceModeler (EVM) v1.1.1 (Haas et al. 2008). For homology-based predictions, a custom database was built by combining the protein sequences from nine insect species downloaded from the Ensembl database release 106 (Agrilus planipennis, Anoplophora glabripennis, Apis mellifera, Dendroctonus ponderosae, Diabrotica virgifera, Drosophila melanogaster, Leptinotarsa decemlineata, Onthophagus taurus, and Tribolium castaneum). Additionally, manually curated insect proteins were retrieved from the UniProtKB/Swiss-Prot release 2022_02 database (UniProt Consortium 2019). To improve genome annotation, we provided transcriptomic data in the form of de novo assembled transcriptomes and Illumina RNA-Seq short reads for M. variabilis and L. trimaculatus (Fratini et al. 2021). These data were processed by the PASA pipeline v2.2.3 (Haas et al. 2003) to reconstruct transcripts and use them as additional evidence provided as input to EVM. The full set of predicted proteins was extracted from the genome of the five species and evaluated for completeness using BUSCO v5.5.0 (Insecta_odb10 database).
Orthology inference and phylogenetic tree construction
Predicted proteins from the five newly assembled blister beetles were analyzed together with genome-predicted proteins from four additional species of Meloidae: Epicauta sibirica (reported as chinensis; Meloinae, Epicautini; JAEDXX000000000); Hycleus phaleratus and H. chicorii (Meloinae, Mylabrini; JACDRP000000000 and JACDRQ000000000, respectively); Meloe dianella (Meloinae, Meloini; JAPTHL000000000) (Fig. 1). We also included the related canthariphilous species Pyrochroa serraticornis (Pyrochroidae; CAJOSL000000000), and Tribolium castaneum (Tenebrionidae; AAJJ00000000). Orthogroups were identified with OrthoFinder v2.4.0 (Emms & Kelly 2019). Mafft v7 (Katoh & Standley 2013) was used to align the protein sequences of each orthogroup. The aligned protein sequences were converted to the corresponding codon sequences using PAL2NAL v14 (Suyama et al. 2006). Phylogeny was inferred using the protein sequences of one-to-one orthologs using rapidNJ (Simonsen et al. 2011) and RAxML v8.2 (Kozlov et al. 2019).
Gene family expansion and contraction
CAFE v5 (Mendes et al. 2020) was used to infer gene birth and death rates (lambda) and retrieve gene families under significant dynamics. As input, we used the species tree with divergence time from the output of MCMCTree and the results of orthogroups from OrthoFinder v2.4.0 (Emms & Kelly 2019). Each orthogroup was deemed to be a gene family. We ran CAFE under the birth–death model to estimate the posterior probabilities of each gene family belonging to different evolutionary rate categories (K). The model of the evolutionary rate categories with the Maximum Likelihood (K = 2) was chosen as the final result. To symbolize each gene family, we took the longest member as representative and BLAST-searched with Diamond (Buchfink et al. 2021) against UniProtKB/Swiss-Prot and NR databases. The best hit from both was retained.
Identification of positively selected genes
The one-to-one orthologs inferred by OrthoFinder v2.4.0 (Emms & Kelly 2019) were used to identify signatures of selection. Positively selected genes were detected with the Codeml program in the PAML package v4.10.6 (Yang 2007). Positive selection signals on genes along specific lineages were detected using the optimized branch-site model following the author's recommendation. A likelihood ratio test (LRT) was conducted to compare a model that allowed sites to be under positive selection on the foreground branch with the null model in which sites could evolve either neutrally or under purifying selection. The P-values were computed based on Chi-square statistics, and genes with P-values less than 0.05 were treated as candidates that underwent positive selection. The web server g:Profiler version e110_eg57_p18_4b54a898 (Kolberg et al. 2023) was used for performing Gene Ontology (GO) (g:GOSt) and pathway enrichment analysis and detecting statistically significantly enriched terms in lineage-specific orthogroups or positively selected genes in Meloidae using D. melanogaster (Flybase) as a reference database for annotation (g:SCS multiple testing correction method, significance threshold of 0.05).
RESULTS
Genome assembly and completeness
The five newly assembled draft genomes of blister beetles ranged from 92.9 (S. caucasica) to 145.4 Mb (M. variabilis) (Table 1), whereas the estimated genome sizes ranged from 86.8 (Z. immaculata) to 186.1 Mb (M. variabilis). The assembled genomes included from 4652 (I. pallidipennis) to 37 634 scaffolds (M. variabilis), with an N50 length spanning 7.0 to 61.8 Kb (Table 1). The gene number (i.e. protein-coding genes) predicted in each genome varied from 12 789 to 17 069, and it translates to a gene density ranging from 117.4 to 142.2 genes/Mb (Table 1), respectively. All genome assemblies received high scores of completeness with BUSCO with values of genes identified as “complete” spanning from 95.3% to 99.0%, and a very low number of core “missing” genes (0.5–1.7%) (Table 1) that indicates that almost all the coding regions were reconstructed in the five genome assemblies. A high level of completeness was also found for the predicted protein-coding gene sets in the five genomes (BUSCO analysis: 90.7–95.6% “complete” genes; 3.1–4.3% “missing” genes). These latter values underscore the efficacy of the annotation process.
| Assembly | Iselma pallidipennis | Zonitis immaculata | Stenodera caucasica | Lydus trimaculatus | Mylabris variabilis |
|---|---|---|---|---|---|
| Estimated genome size (Mb) | 101.7 | 86.8 | 105.6 | 104.3 | 186.1 |
| Assembled genome size (Mb) | 107 | 102 | 92.9 | 114.1 | 145.4 |
| Scaffold N50 (Kb) | 61.8 | 15.9 | 31.2 | 45.2 | 7 |
| Scaffold number | 4652 | 16 196 | 12 385 | 8865 | 37 634 |
| Repeat content (%) | 8.82 | 11.10 | 9.19 | 9.21 | 8.28 |
| Protein coding genes | 13 171 | 12 789 | 13 201 | 13 596 | 17 069 |
| Gene density (Mb) | 123.1 | 125.4 | 142.2 | 119.2 | 117.4 |
| BUSCO | |||||
| Complete | 1003 (99.0%) | 983 (97.0%) | 986 (97.3%) | 991 (97.8%) | 965 (95.3%) |
| Complete and single copy (S) | 1000 | 981 | 983 | 989 | 964 |
| Complete and duplicated (D) | 3 | 2 | 3 | 2 | 1 |
| Fragmented (F) | 5 | 20 | 18 | 12 | 30 |
| Missing (M) | 5 | 10 | 9 | 10 | 18 |
Identification of ortholog genes and gene family expansion and contraction
Overall, we identified 14 063 orthogroups (Table S1, Supporting Information). Among them, 37 (0.2%) were found only in blister beetles, whereas 228 (1.6%) were exclusive to P. serraticornis (Fig. 2) and mostly associated with monooxygenase activity (GO0004497) (Table S9, Supporting Information). 117 (0.8%) orthogroups were shared between Meloidae and Pyrochroidae, with the SNARE vesicle transport pathway (Kyoto Encyclopedia of Genes and Genomes, KEGG:04130) significantly over-represented (Table S9, Supporting Information). In each lineage, we recorded specific orthogroups related to chemosensory perception (odorant, ionotropic, and gustatory receptors or odorant binding proteins) and/or detoxification (e.g. cytochrome P450) (Table S1, Supporting Information).
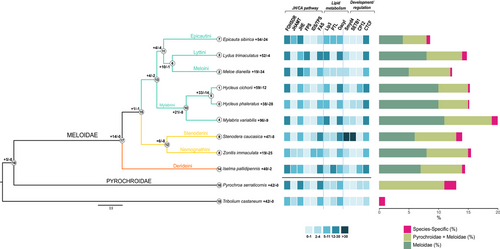
The phylogenetic tree built using the single-copy ortholog genes (Fig. 2) was consistent with previous phylogenetic studies on the family (Riccieri et al. 2022; Riccieri et al. 2023). Meloidae was monophyletic and sister to P. serraticornis, and the three subfamilies were distinct: Eleticinae (i.e. I. pallidipennis) was sister to Nemognathinae and Meloinae. Within this latter subfamily, Mylabrini was monophyletic and separated from Epicautini, Meloini, and Lyttini (Fig. 2).
Five gene families expanded in Meloidae + Pyrochroidae (Node: 19, Fig. 2; Table S2, Supporting Information), among which we detected orthologs of the farnesol dehydrogenase (FOHSDR) involved in insect hormone biosynthesis (e.g. juvenile hormone pathway) and cytochromes (i.e. CYP450 6a23) responsible for the oxidation/degradation of xenobiotics and/or insect hormones. Along this lineage, genes responsible for de novo biosynthesis of long-chain saturated fatty acids (e.g. fatty acid synthases) expanded as well. We also observed the expansion of protein families involved in trans-membrane transportation/secretion of (modified) compounds, such as the ATP-binding cassette (ABC) transporters subfamily C member 4 transporters and proteins containing the Sec14p-like lipid-binding domain (e.g. over-represented with 26 copies in P. serraticornis; Table S2, Supporting Information). 14 gene families expanded specifically in Meloidae (Node: 17, Fig. 2; Table S2, Supporting Information). CYP450 member 6 (i.e. 6a14 and 6k1) or other CYP450 families and monooxygenases (e.g. luciferin 4) responsible for the metabolism of xenobiotics and/or insect hormones largely duplicated in this lineage (but also independently—multiple times—in some of the examined blister beetle species). Key enzymes of the juvenile hormone (JH) pathway significantly expanded in Meloidae genomes: Juvenile hormone acid O-methyltransferase (JHAMT), FOHSDR, farnesyl pyrophosphate synthase (FPS), and juvenile hormone esterase (JHE, degrading JH) underwent significant expansions in I. pallidipennis (n = 5 copies), E. sibirica (n = 21 copies), L. trimaculatus (n = 20 copies), and M. dianella (n = 22 copies), respectively (Fig. 2; Table S2, Supporting Information). Also, protein families involved in both metabolite transport (ABC member 4) and uptake of chemicals (e.g. from the intestine to surrounding tissues, including gonads, such as nrf-6 that counts 14 copies in S. caucasica) expanded in Meloidae, as well as proteins for odorant perception. Of note, the largest duplication of odorant receptors occurred independently in I. pallidipennis (n = 81 copies of putative Or4) and L. trimaculatus (n = 28 copies of putative Or4) (Table S2, Supporting Information). In I. pallidipennis (tip: 14, Fig. 2; Table S2, Supporting Information), we detected a relatively large expansion of enzymes (CYP450 6a23; ABC member 4) involved in xenobiotic and/or insect hormone metabolism/transport. Other protein families that duplicated in this lineage were involved in fatty acid metabolism/mobilization (i.e. fatty acid synthase and lipases; Fig. 2), immunity (e.g. phenoloxidase-activating factor 2), regulatory functions (e.g. various repressors), and development (e.g. flexible cuticle 12-like protein; Fig. 2). Six families specifically expanded in Nemognathinae (Node: 12, Fig. 2; Table S2, Supporting Information), such as terpene/isoprenoid synthases (IDSs/TPSs) (n = 8 copies in Z. immaculata), digestive cysteine proteinases, and proteins involved in histone methylation/epigenetic control (e.g. Pre-SET, SET, and MYND domains were over-represented in S. caucasica), whereas the four expanded families in Meloinae (Node: 13, Fig. 2; Table S2, Supporting Information) were mainly related to digestion and lipid metabolism (e.g. transmembrane protease serine 9 and lipase 3; Fig. 2).
We also observed the expansion of detoxification enzymes—other than CYP450—in specific Meloidae lineages (i.e. in L. trimaculatus and M. variabilis), such as aldo–keto reductases, UDP-glycosyltransferases, and carboxylic ester hydrolases (Table S2, Supporting Information). Other genes were gained in various blister beetle species to perform fatty acid modification (e.g. O-acyltransferase-like proteins in M. variabilis; Fig. 2) or reception/transport of glutamate (e.g. vesicular glutamate transporter 3 in E. sibirica; glutamate receptor in S. caucasica) (Table S2, Supporting Information). Finally, collagenases—possibly involved in wound/damage healing—were duplicated in M. dianella (i.e. collagenase 3), as well as venom components/allergens common in other organisms in E. sibirica (venom serine carboxypeptidase) and S. caucasica (venom acid phosphatase Acph-1) (Table S2, Supporting Information).
Identification of positively selected genes
152 genes underwent positive selection in Meloidae + Pyrochroidae (Table S3, Supporting Information), with an over-representation of biological processes related to nucleus maintenance (GO1900182 and GO0017056) and cell–cell adhesion (GO0044331) (Table 2; Table S9, Supporting Information). The nucleocytoplasmic transport pathway (KEGG:03013) was also significantly represented (Table 2; Table S9, Supporting Information). Several positively selected genes were engaged in lipid modification and transport (e.g. glycerol-3-phosphate phosphatase, sphingosine kinase, and long-chain fatty acid transport protein 1), as well as in vesicle formation/mobilization or exo–endocytosis (e.g. AP2-associated protein kinase 1 regulating clathrin-mediated endocytosis, striatin-interacting protein 1, low-density lipoprotein receptor-related protein 1). We also detected positively selected orthologs of proclotting enzymes—for example, converting coagulogen to insoluble coagulin gel—or genes involved in mechanical sensing (i.e. protein-stum like) and muscle contraction/relaxation (i.e. twitchin and calponin). These latter (out of the 37 genes scored) were also specifically recorded in Meloidae (Table S4, Supporting Information), as well as several genes involved in phase I xenobiotic degradation (e.g. carboxylesterase CXE18) and transport (e.g. msta and NPC intracellular cholesterol transporter 1). In Nemognathinae, most of the 234 positively selected genes (Table S5, Supporting Information) were devoted to gene regulation, mainly for transcriptional repression (e.g. Polycomb group proteins involved in methylation), DNA/RNA binding or processing, or, more specifically, to assist development (GO terms such as GO0048513 “animal organ development” and GO0007275 “multicellular organism development,” and KEGG terms as KEGG:04320 “Dorso-ventral axis formation” and KEGG:043320 “Notch signaling pathway” were, enriched; Table 2; Table S9, Supporting Information). Many other genes in this subfamily that were involved in cellular processes (e.g. GO0009987, Table 2) resulted positively selected, such as those promoting vesicular formation/secretion (e.g. exocyst complex component Epsin-1) and intracellular trafficking (e.g. sorting nexin-4 and GTPases Rab) (Table S5, Supporting Information). Others were implied in the transport of compounds (e.g. monocarboxylate transporter 10) and lipid conversion and metabolism (e.g. diacylglycerol lipase-alpha isoform X2) (Table S5, Supporting Information). In Mylabrini (Table S6, Supporting Information), most of the 323 selected genes were for DNA/RNA regulatory functions (e.g. regulation of transcription and RNA metabolism; GO0045944, GO0051253), capable of responding to abiotic stimuli (e.g. GO0009628) or related to development and morphogenesis (e.g. embryo development GO0009790; regionalization GO0003002) (Table 2; Table S9, Supporting Information). Of note, the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) of the mevalonate (MVA) pathway (leading to the production of non-sterol isoprenoids) was positively selected in this lineage. Most of the remaining genes in Mylabrini were involved in lipid metabolism/transport (e.g. MFS1) or in vesicular trafficking (e.g. sorting-nexin-4; clathrin heavy chain; calcineurin B) (Table S6, Supporting Information). In Lyttini + Meloini + Epicautini, the 37 positively selected genes (Table S7, Supporting Information) were mostly involved in biological processes such as “cell junction assembly” (GO0034329) and “regulation of anatomical structure size” (GO0090066) (Table 2; Table S9, Supporting Information). Among all, we detected genes contributing to vesicle formation and movement (e.g. Sec23 trunk, Cadherin 86D, and Rab4/rab5 effectors), development (e.g. neurexin-4), and epigenetic control (e.g. mst-A and mst-B). 270 genes were positively selected in I. pallidipennis (Table S8, Supporting Information) and were mostly devoted to cell development, differentiation (or in general to developmental processes; GO0048468, GO0048869, and GO0030154), and anatomical structure morphogenesis (GO0009653) (Table 2; Table S9, Supporting Information).
| Lineage | Term id | Term description | Adj. P | Term id | Term description | Adj. P |
|---|---|---|---|---|---|---|
| Meloidae + Pyrochroidae | GO:0017056 | Structural constituent of nuclear pore | 0.027 | GO:0044 331 | Cell–cell adhesion mediated by cadherin | 0.035 |
| GO:1900182 | Pos. reg. of protein localization to nucleus | 0.003 | KEGG:03 013 | Nucleocytoplasmic transport | 0.019 | |
| Nemognathinae | GO:0048513 | Animal organ development | 0.002 | KEGG:04 330 | Notch signaling pathway | 0.022 |
| GO:0007275 | Multicellular organism development | 0.047 | KEGG:04 320 | Dorso-ventral axis formation | 0.035 | |
| Mylabrini | GO:0009987 | Cellular process | 2.001 × 10−6 | GO:0032 989 | Cellular component morphogenesis | 0.012 |
| GO:0045944 | Pos. reg. of transcription by RNA pol II | 0.016 | GO:0032 502 | Developmental process | 4.432 × 10−7 | |
| GO:0009628 | Response to abiotic stimulus | 0.036 | GO:0035 282 | Segmentation | 0.006 | |
| GO:0051253 | Neg. reg. of RNA metabolic process | 0.044 | GO:0009 790 | Embryo development | 6.378 × 10−7 | |
| GO:0009653 | Anatomical structure morphogenesis | 3.665 × 10−10 | GO:0007 399 | Nervous system development | 2.971 × 10−4 | |
| GO:0003002 | Regionalization | 1.072 × 10−7 | GO:0032 501 | Multicellular organismal process | 3.684 × 10−8 | |
| GO:0007350 | Blastoderm segmentation | 0.011 | KEGG:04 330 | Notch signaling pathway | 0.003 | |
| Lydus + Meloe + Epicauta | GO:0034329 | Cell junction assembly | 0.031 | GO:0090 066 | Reg. of anatomical structure size | 0.038 |
| Iselma | GO:0048468 | Cell development | 1.170 × 10−5 | GO:0048 869 | Cellular developmental process | 3.553 × 10−5 |
| GO:0030154 | Cell differentiation | 3.509 × 10−5 | GO:0009 653 | Anatomical structure morphogenesis | 2.842 × 10−4 |
DISCUSSION
All Meloidae endogenously produce CA to be used as a defensive compound through reflex bleeding (Carrel & Eisner 1974). With the possible exception of Eleticinae, blister beetles undergo a hypermetamorphic larval development (Bologna et al. 2001, 2010). Anyhow, this beetle family shows a wide array of species-specific ecological adaptations at the larval stage, related to dispersion (e.g. phoresy) and host selection (Bologna et al. 2010; Bologna & Di Giulio 2011).
We examined from an evolutionary perspective the molecular basis of common features across the three Meloidae subfamilies and of peculiar adaptations in specific blister beetle lineages, by comparing five newly generated genomes with those already available for this beetle family (Fig. 1). We also included genomic data from the red-headed cardinal beetle P. serraticornis, a closely-related canthariphilous species (Cai et al. 2022), to unveil common genomic traits possibly related to CA metabolism.
We generated five novel draft genome assemblies of high quality, as suggested by the high percentage of (BUSCO) completeness in terms of the expected gene content (Table 1). The overall sizes of these newly assembled genomes (<90–145 Mb) were almost similar and comparable with those already scored for other Meloidae (Tian et al. 2021; Zhou et al. 2023) (Table 1). In general, the estimated genome sizes of Hycleus and Mylabris were confirmed to be larger than those of all other species, apparently due to a relatively higher gene number content (Table 1). The already observed expansion of chemosensory receptor gene families in Mylabrini (Wu et al. 2020), as also suggested by our findings (e.g. OG0001043, GR92; OG0000103, OBP18), probably contributed to inflating their genome sizes and might have represented the key to adaptive radiation in Hycleus, the most diverse genus of Meloidae, counting more than 500 species (Riccieri et al. 2020; Wu et al. 2020).
Several proteins devoted to the mobilization of compounds and/or related to vesicular formation and solute transport were overrepresented and/or positively selected in both Meloidae and Pyrochroidae (Tables S2,S3,S9, Supporting Information). Within Meloidae, in particular, proteins conferring these functions underwent positive selection in Nemognathinae and Mylabrini (Tables S2,S5, Supporting Information). This is consistent with the hypothesis that CA could be sequestered in intracellular compartments to prevent its toxicity within the bodies of both insect groups (Fratini et al. 2021; Muzzi et al. 2022; Molfini et al. 2023). Indeed, numerous replenished vesicles were found to populate the cells of glandular epithelia of male reproductive organs of Meloe proscarabaeus (Muzzi et al. 2022) and of the cranial apparatus of Pyrochroa coccinea (Molfini et al. 2023). Moreover, ABC transporters and other protein families involved in solute mobilization were found over-expressed in males of Meloidae (Fratini et al. 2021; Zhou et al. 2023). These were hypothesized to be engaged in the compartmentalization of toxic substances, such as CA, into vesicles, to ensure protection of the male reproductive tract (Fratini et al. 2021). Other orthologs positively selected in Meloidae + Pyrochroidae were involved in different physiological functions, such as muscle contraction/relaxation, mechanosensory transduction, and coagulation (Table S3, Supporting Information). Genes performing these functions were plausibly under adaptive selection in both taxa to perform the emission of CA in response to external stimuli and possibly repair damaged tissues after autohemorrhaging (e.g. through a purported collagenase activity in M. dianella) or glandular secretion (Molfini et al. 2023). Consistently, previous transcriptome analyses revealed a compelling set of genes (moderately to highly) expressed in M. variabilis and L. trimaculatus exhibiting homology with the bovine pancreatic trypsin inhibitors (BPTI) and Kunitz-type protease inhibitors (KTPI) of A. glabripennis (Fratini et al. 2022). BPTI and KTPI are known to possess potent anticoagulant activities (García-Fernández et al. 2016). Hence, the 33 transcripts retrieved in these two species were hypothesized to provide an efficient regulation of coagulation to minimize hemolymph loss during the autohemorrhaging process (Fratini et al. 2022). Overall, protein families involved in fatty acid biosynthesis underwent an expansion in both Meloidae and Pyrochroidae and also specifically along most blister beetle lineages (Fig. 2; Table S2, Supporting Information). Lipids play multiple roles in insect metabolism and development, such as energy storage, signal transduction, cell membrane formation, and compound synthesis (Arrese & Soulage 2010; Song et al. 2022). In particular, in Meloidae, the high number of genes devoted to lipid metabolism—also noted before (Tian et al. 2021)—could prove a sustained activity of fat bodies, also with its purported crucial role in CA production (Jiang et al. 2019; Fratini et al. 2021). Fatty acids in insects also serve as precursors in the synthesis of eicosanoids and pheromones, and hence their contribution to CA biogenesis (e.g. precursors) cannot be excluded (Jiang et al. 2019; Fratini et al. 2021). Protein families implied in the transport of lipids and/or apolar molecules also experienced gene duplication and positive selection in various species of blister beetles and in P. serraticornis (Table S2, Supporting Information). Of note, numerous copies of nrf-6-like proteins—known to mediate the transport of xenobiotics from the intestine to the surrounding tissues, including the reproductive tract (Harrison et al. 2015)—were found in Meloidae. These lipid/xenobiotic transport proteins could vehiculate CA and mitigate its reactivity while circulating in beetle tissues, as previously suggested for lipocalins (e.g. ApoD) that were found abundantly expressed in the hemolymph of Meloidae (Fratini et al. 2021). CYP450s expanded as well in both Meloidae and Pyrochroidae (Table S2, Supporting Information). Given their recognized role in detoxification and ubiquitous expression in various insect tissues (Scott et al. 1998; Scott & Wen 2001), CYP450s can prevent the detrimental effects of chemical plant xenobiotics after ingestion in members of both the examined beetle families. We might also suggest that the cysteine–proteinases specifically expanded in Nemognatinae (Table S2, Supporting Information) are likely to assist in similar digestive functions (Chen et al. 2013). However, CYP450s also have roles in pheromone and endogenous compound biosynthesis (Scott & Wen 2001). In particular, CYP6 and other monooxygenases (e.g. luciferase-like) that specifically (and independently) expanded in Meloidae might take part in the metabolism of hormones, as in other insects (Scott & Wen 2001). Similarly, aldo-keto reductases, UDP-glycosyltransferases, and carboxylic ester hydrolases, specifically expanded in Meloinae, could also have a role in the biotransformation of endogenous chemicals (Barski et al. 2008; Ahn et al. 2012; Cruse et al. 2023). Since CA is a by-product of the JH, we cannot rule out that some CYP450s (and/or the above-mentioned enzyme classes) may function in a not-yet-clarified JH degradation step crucial for CA biosynthesis (but see also Tian et al. 2021; Zhou et al. 2023). JH is synthesized via the MVA pathway, with farnesol as an intermediate (see Fratini et al. 2021). The need to supply large amounts of this hormonal substrate to produce CA likely led blister beetle genomes to evolve multiple copies of key enzymes taking part in the upstream terpenoid backbone biosynthesis (i.e. HMGCR, IDS/TPS, and FPS) and downstream JH pathway (i.e. FOHSDR, converting farnesol into farnesal; JHAMT, converting JH acid into JH III; JHE, converting JH diol into JH acid diol, a purported cantharidin precursor; see Jiang et al. 2019; Wu et al. 2023) (Fig. 2).
Our data indicate that genes devoted to the regulation of development and morphogenesis significantly evolved in Meloidae (Table 2). This is not unexpected given the hypermetamorphic development of most members of this family, composed of larval instars showing specific phenotypes and playing different roles in blister beetle development (Bologna et al. 2010; Bologna & Di Giulio 2011). Specifically, the first instar (triungulin) is devoted to dispersal and host/food-source finding, while the subsequent larval stages are responsible for feeding and body growth (first grub), enduring periods of cold or dryness (coarctate), and preparing the pupal chamber (second grub) (Bologna et al. 2010; Bologna & Di Giulio 2011). It has been suggested that environmental conditions and larval diets may alter the expression of proteins related to carbohydrate and energy metabolism, immunity, digestion, and absorption of nutrients during development (Li et al. 2014). Coherently, the expansion of homologous genes of, for example, SET and MYND domain-containing proteins (SMYD4; Fig. 2), a special class of protein lysine methyltransferases involved in methylation of histones and non-histone targets (Spellmon et al. 2015), might have evolved to regulate the fine-tuned gene expression required according to larval stages. Hence, the epigenetic control might be also functional to express genes needed to perform the different ecological adaptations observed in larval habits of Meloidae species, such as phoresy or crawling and specialization to a preferred host (i.e. wild bees or grasshoppers; Fig. 1) (Bologna et al. 2010; Bologna & Di Giulio 2011).
In conclusion, most of the major adaptive genetic traits observed in blister beetles can be interpreted in light of CA production and hypermetamorphosis, which certainly represent key features that emerged during the evolutionary history of Meloidae. Since JH maintains juvenile phenotypes by preventing the metamorphosis induced by the molting hormone ecdysone (Yokoi et al. 2020), the gain of copies of enzymes involved in JH biosynthesis in Meloidae might have been functional to enhance both CA production and the prolonged larval phases characterizing the hypermetamorphic development (see also Li et al. 2014). Hence, we cannot exclude that still-unveiled pleiotropic effects by some genetic traits might regulate these two crucial aspects of blister beetle biology and have concurred to their evolutionary success.
ACKNOWLEDGMENTS
This research was funded by the project “NOCLOT—NuOvi farmaCi anticoaguLanti dalla biOdiversiTà dei meloidi” financed by Regione Lazio (grant nos. A0375-2020-36555 and CUP F85F21003680009), and co-funded by MIUR-Italy Grants of Departments of Excellence—L. 232/2016—art.1 cc. 314–337 awarded to the Department of Science of Roma Tre University (2018–2022 and 2023–2027). A.R. is currently supported by the project PON—Ricerca e Innovazione (MUR; Project Code: 999900_PON_RTD_A7-G-15023_SCIENZE). M.A.B. and E.M. acknowledge the support of NBFC to the University of Roma Tre—Department of Science and Sapienza University—Department of Biology and Biotechnologies, “Charles Darwin”, funded by the Italian Ministry of University and Research, PNRR, Missione 4 Componente 2, “Dalla ricerca all'impresa”, Investimento 1.4 (Project CN00000033). The project was partly supported also by Rome Technopole, PNRR grant M-4C-2Inv. 1.5 CUP F832B22000040006 to MAB.




