Biodiversity of Metarhizium From Korea and Pathogenicity Screening of Two Unrecorded Species Metarhizium lepidiotae and Metarhizium robertsii
Funding: This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry, Agricultural Machinery/Equipment Localization Technology Development Program, and Ministry of Agriculture, Food and Rural Affairs (321054-05).
ABSTRACT
This study analyzed the biodiversity of entomopathogenic fungi Metarhizium species isolated from Korean soil samples and insect cadavers. Morphological characteristics and molecular phylogenetic analyses were used to analyze Korean species diversity. Concatenated datasets of three genes, the translation elongation factor 1-a (TEF), DNA-directed RNA polymerase II (RPB1), and internal transcribed spacer (ITS), were used for phylogenetic analyses using the following three methods: maximum parsimony (MP), Bayesian inference (BI), and maximum likelihood (ML). In addition, the TEF gene was used for molecular identification. As a result, five species were identified: Metarhizium pinghaense, Metarhizium robertsii, Metarhizium rileyi, Metarhizium pemphigi, and Metarhizium lepidiotae. In particular, M. robertsii and M. lepidiotae were identified in Korea for the first time in this study. The TEF gene was useful for species identification of Metarhizium species especially M. pinghaense and Metarhizium anisopliae. Among these fungi, only M. lepidiotae showed high pathogenicity against cotton aphids (Aphis gossypii) and lesser mealworm (Alphitobius diaperinus).
1 Introduction
The genus Metarhizium is a widespread genus of entomopathogenic fungi and is known for its diverse asexual reproductive morphologies and life stages (Bischoff, Rehner, and Humber 2009; Kepler et al. 2012; Mongkolsamrit et al. 2020). Most species produce greenish conidia on the cadaver of arthropod hosts and are well known as “green muscardine fungus” (Roberts and St. Leger 2004). This genus Metarhizium was first described by Sorokin 1879 based on Metarhizium anisopliae (Metchnikoff 1879) (Hypocreales: Clavicipitaceae) and is one of the most notable entomopathogenic fungi (Shin et al. 2013). Most of the species have been isolated from soil or close association with plant roots, sometimes found on Arthropoda cadavers with conidia (Hu and St Leger 2002; Wyrebek et al. 2011; Nishi and Sato 2017).
M. anisopliae is one of the most important groups of entomopathogenic fungi and Beauveria bassiana (Bals.-Criv.) Vuill., 1912 (Hypocreales: Clavicipitaceae), for commercially developed microbial pesticides. Before the year 2000, the taxonomy of Metarhizium focused on morphological characteristics, such as the shapes of the conidia and phialides. Since 2000, molecular analyses have utilized the internal transcribed spacer (ITS) region and random amplified polymorphic DNA (RAPD) pattern (Driver, Milner, and Trueman 2000). Multigene phylogenetic analyses elevated various Metarhizium varieties to species level in the M. anisopliae and Metarhizium flavoviride lineages (Bischoff, Rehner, and Humber 2006, 2009). Currently, the multiphylogenetic analysis method is widely used when securing the diversity of many Metarhizium species and in phylogenetic relationships.
Recently, 101 species of Metarhizium have been reported worldwide (Bischoff, Rehner, and Humber 2009; Lopes et al. 2018; Mongkolsamrit et al. 2020; Species of Fungorum, 2024). However, only six species have been recorded in the genus Metarhizium species in Korea (Table 1). Moreover, most of the studies on Metarhizium in Korea have focused on biological control or described the species shown in the screening of entomopathogenic fungi, and there have been poor taxonomic studies focused on the genus of Metarhizium.
| Species | Strains | Source | Location | Reference |
|---|---|---|---|---|
|
Metarhizium anisopliae Metarhizium pinghaense |
J-22 3-1-2 |
Soil Soil |
Jeju Island Korea |
Lee et al. 1997 Ko et al. 2021 |
| Metarhizium pinghaense | 4-4-1 | Soil | Korea | Ko et al. 2021 |
| M. anisopliae | FT83 | Soil | Yangpyeong-gun, GG | Han, Kim, and Lee 2014 |
| M. koreanum | BPI892885 | Planthoppers | Korea | Kepler et al. 2014 |
| M. marquandii | DUCC5194 | — | Cheongyang-gun, CN | Ahn et al. 2018 |
| M. pemphigi | CNUFC AS1-26 | Soil | Taean-gun, CN | Pangging, Nguyen, and Lee 2021 |
| M. rileyi | KNU-Gunwi 2B | Soil | Gunwi-gun, GB | Moe et al. 2021 |
| M. majus | GK1 | Coleoptera | Gimhae-si, GN | Kwak et al. 2021 |
| M. majus | GK2 | Coleoptera | Gimhae-si, GN | Kwak et al. 2021 |
- Abbreviations: CN, Chungcheongnam-do; GB, Gyeongsangbuk-do; GG, Gyeonggi-do; GN, Gyeongsangnam-do.
Therefore, this study attempted to determine the species diversity in the Korean genus Metarhizium based on the morphological characteristics and phylogenetic relationship of the entomopathogenic fungi collected from cadavers and soil samples. Additionally, we identified Korean Metarhizium strains and evaluated their potential for pest control. This data provides a foundation for developing biopesticides targeting agricultural insect pests such as Aphis gossypii Glover, 1877 (Hemiptera: Aphididae), common name cotton aphids, and Alphitobius diaperinus Panzer, 1797 (Coleoptera: Tenebrionidae), common name lesser mealworm, which are significant pests in agricultural environments and animal husbandry.
2 Materials and Methods
2.1 Fungal Isolates
The genus Metarhizium isolates in this study were collected from forest soil samples and insect cadavers. The isolates from the soil samples were isolated using the insect bait method (Kim et al. 2018), and the isolates from the cadavers were isolated by spreading the conidia formed on the cadavers onto a PDA medium (Shin et al. 2011). Cultured fungi were reisolated on PDA plates several times to obtain pure cultures. The isolates used in this study are listed in Table 2.
| Identification | Isolates | Source | Location | Reference |
|---|---|---|---|---|
| M. lepidiotae | KW9R2W1 | Soil | Hoengseong-gun, GW | This study |
| M. pemphigi | IPBL-H | Soil | Gwangju, Buk-gu, JN | This study |
| M. pinghaense | IPBL-I | Soil | Gwangju, Buk-gu, JN | This study |
| M. rileyi | IPBL-J | Lepidoptera | 150 Cheonseo-gil, Chunpo-myeon, Iksan-si, Jeollabuk-do, JB | This study |
| M. robertsii | KB6T1W1 | Soil | Yeongju-si, GB | This study |
- Abbreviations: GB, Gyeongsangbuk-do; GW, Gangwon-do; JB, Jeollabuk-do; JN, Jeollanam-do.
2.2 Morphological Identification
Single colony purified strains were inoculated in the three positions of PDA plates and allowed to grow. Conidia and growth were examined under a dissecting microscope (Olympus SZX16). For characterization, conidia measurements and imaging were conducted using a Zeiss Axio Imager A2 (Carl Zeiss, Germany) compound microscope equipped with an Optinity KCS2000SS digital camera (Korealabtech, Korea).
Cultures were grown on PDA (Difco) and SDA (Difco) (Bischoff, Rehner, and Humber 2009) and incubated at 25°C in dark conditions. Sporulation was followed by triple colony culture method and observed on the two types of media. Additionally, a single colony was also cultured for 10 days for observation of the basal side. Images after 14 days of cultures on the two media were collected. The diameter range of each colony was measured, and images of growth on PDA were captured. Conidial length and width were measured from 20 individual conidia.
2.3 DNA Extraction
Fungi were placed in 1.5-mL microcentrifuge tube with 650 μL of fungal DNA extraction buffer (0.2 M Tris-Cl (pH 7.5), 0.5 M NaCl, 10 mM EDTA (pH 8.0), and 1% (w/v) SDS). After grinding, 650 μL of phenol–chloroform–isoamyl alcohol (25:24:1) was added to each tube. The mixture was vortexed for 15 min at room temperature (RT). After vortexing, the sample was centrifuged at 12,300 × g for 10 min at RT. The supernatant was transferred to a new 1.5-mL microcentrifuge tube. To precipitate the DNA, 1 mL of 100% ethanol was added, followed by gentle vortexing. The mixture was then centrifuged at 25,900 × g for 15 min. The supernatant was discarded, and the pellet was washed with 1 mL of 70% ethanol followed by drying in RT for 15 min. After drying, 50 μL of prewarmed ddH₂O (40°C) was added to dissolve the DNA by gentle tapping. The extracted DNA was stored at −20°C and later used as a template for PCR amplification.
2.4 PCR Amplification and Sequencing
The genes used in this study were translation elongation factor 1-alpha (TEF-1a), the largest subunits of DNA-directed RNA polymerase (RPB1), and ITS. Primer sets used in the experiments are listed in Table S1. The PCR reaction mixture consisted of GenomicsOne 5x PCR premix (GenomicsOne Co.), 100 ng DNA, and 10 pmoL of each primer in a 20 μL volume. The reaction parameters were as follows: initial denaturation for 2 min at 94°C; followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final 5-min extension at 72°C using a thermal cycler (Takara, Japan). After PCR, the amplified DNA fragments were separated using electrophoresis in 1.0% agarose gel and sequenced directly (Macrogen Co.) The nucleotide sequences from isolated isolates were submitted to the GenBank database (Table 3).
| Species | Strains | Locality | Host | tef | rpb1 | ITS | Reference |
|---|---|---|---|---|---|---|---|
| Beauveria bassiana | 42R | Korea | Myzus persicae | ON479087 | ON479095 | ON406170 | This study |
| M. acridum | ARSEF 7486 | Niger | Orthoptera | EU248845 | EU248897 | HQ331458 | Bischoff, Rehner, and Humber (2009) |
| M. album | ARSEF 2082 | Indonesia | Hemiptera | DQ522352 | DQ522398 | AY375446 | Sung et al. (2007) |
| M. biotecense | BCC51812 | Thailand | Hemiptera: Delphacidae | MN781693 | MN781745 | MN781878 | Mongkolsamrit et al. 2020 |
| M. eburneum | BCC79252 | Thailand | Lepidoptera pupa | MN781682 | MN781736 | MN781914 | Mongkolsamrit et al. 2020 |
| M. flavoviride | CBS 125.65 | United States | — | MT078846 | MT078862 | MT078885 | Mongkolsamrit et al. 2020 |
| M. frigidum | ARSEF 4124 | Australia | Coleoptera | DQ464002 | DQ468361 | NR132012 | Bischoff, Rehner, and Humber (2006) |
| M. fusoideum | BCC28246 | Thailand | Lepidoptera | MN781699 | MN781749 | MN781893 | Mongkolsamrit et al. 2020 |
| M. globosum | ARSEF 2596 | India | Lepidoptera | EU248846 | EU248898 | NR132020 | Bischoff, Rehner, and Humber (2009) |
| M. guizhouense | CBS 258.90 | China | Lepidoptera | EU248862 | EU248914 | HQ331448 | Bischoff, Rehner, and Humber (2009) |
| M. koreanum | BCC30455 | Thailand | Hemiptera: Fulgoromorpha | MN781701 | MN781750 | MN781904 | Mongkolsamrit et al. 2020 |
| M. lepidiotae | ARSEF 7488 | Australia | Coleoptera | EU248865 | EU248917 | HQ331456 | Bischoff, Rehner, and Humber (2009) |
| M. lepidiotae | KW9R2W1 | Korea | Hemiptera | ON479088 | ON479096 | ON406200 | This study |
| M. majus | ARSEF 1015 | Japan | Lepidoptera | EU248866 | EU248918 | HQ331444 | Bischoff, Rehner, and Humber (2009) |
| M. majus | ARSEF 1914 | Philippines | Coleoptera | EU248868 | EU248920 | HQ331445 | Bischoff, Rehner, and Humber (2009) |
| M. minus | ARSEF 2037 | Philippines | Hemiptera | DQ522353 | DQ522400 | AF138271 | Sung et al. (2007) |
| M. pemphigi | IPBL-H | Korea | Coleoptera | ON479089 | ON479097 | ON406201 | This study |
| M. phasmatodeae | BCC2841 | Thailand | Phasmatodea | MN781681 | MN781738 | MN781911 | Mongkolsamrit et al. 2020 |
| M. pinghaense | CBS 257.90 | China | Coleoptera | EU248850 | EU248902 | HQ331450 | Bischoff, Rehner, and Humber (2009) |
| M. pinghaense | IPBL-I | Korea | Coleoptera | ON479090 | ON479098 | ON406203 | This study |
| M. rileyi | CBS 806.71 | United States | Trichoplusia ni (Noctuidae) | EF468787 | EF468893 | AY624205 | Sung et al. (2007) |
| M. rileyi | IPBL-J | Korea | Lepidoptera: Noctuidae | ON479086 | ON479093 | ON406204 | This study |
| M. robertsii | KB6T1W1 | Korea | Coleoptera | ON479091 | ON479099 | ON406205 | This study |
2.5 Molecular Phylogenetic Analyses
The DNA sequences generated in this study were assembled and edited using Geneious Prime 2022 (Version 1.1). Multiple sequence alignments of TEF, RPB1, and ITS were carried out using MAFFT v7.490 (Katoh and Standley 2013). The phylogenetic analyses of TEF and the combined dataset of the three genes (TEF + RPB1 + ITS) were analyzed using the Bayesian inference (BI), maximum parsimony (MP), and maximum likelihood (ML) methods. For ML analysis in MEGA X (Stamatakis 2014), the General Time Reversible model was used (GTR + G + I), and the supports were estimated using the bootstrap values obtained in maximum likelihood (MLBS) with 2000 replicates. The MP analysis was conducted on the combined dataset using the same program. Bootstrap values obtained in maximum parsimony (MPBS) analysis was performed using the maximum parsimony criterion in 1000 replications. For BI analysis, the GTR substitution model was used with the MrBayes plugin in Geneious Prime Version 2022.1.1, with a 1,100,000 chain length and a 100,000 burn-in length. Interactive Tree Of Life (iTOL) v4.2 was used for tree visualization. The sequences of TEF, RPB1, and ITS of 15 representative Metarhizium species for the phylogenetic tree analysis of this study are listed in Table 3.
2.6 Conidial Preparation
All fungal strains were cultured on PDA at 25 ± 1°C for 14 days. Conidia were collected by suspending the fungal cultures in sterile distilled water containing 0.05% (v/v) Tween 80 (Sigma-Aldrich). The suspension was filtered to remove mycelial debris, and the conidial concentration was adjusted to 1 × 108 conidia/mL using a hemocytometer.
2.7 Bioassays Against Cotton Aphids and Lesser Mealworms
Cotton aphids (A. gossypii) were reared on potted cucumber plants (Cucumis sativus L., 1753) in Plexiglas cages at 25 ± 1°C under a photoperiod of 16:8 h (L:D) in an insect rearing room. To obtain first-instar nymphs, adult aphids were placed on 30-mm cucumber leaf discs in 35 mm Petri dishes containing 1.5% agar. After 24 h, the adults were removed, leaving behind the newly emerged nymphs. Each leaf disc with 10 nymphs was prepared for the bioassay.
Lesser mealworms (A. diaperinus) were reared and maintained in Plexiglas cages at 25 ± 1°C, with a photoperiod of 16:8 h (L:D) in an insect rearing room. The 10 larvae of lesser mealworms were placed on 55-mm filter paper in a 60-mm diameter insect breeding dish (SPL co.) and used for bioassay.
Conidia were harvested as previously described above, and a 1-mL aliquot of each conidial suspension was sprayed onto the breeding dish from a vertical height of 30 cm using an airbrush. After treatment, the samples were incubated in a Plexiglass cage at 25 ± 1°C with > 70% relative humidity (RH) and a 16:8 h (L:D) photoperiod to observe the mortality for 7 days at 24 h intervals. The 0.05% Tween 80 solution was used as a negative control, and all bioassay experiments were repeated three times.
2.8 Statistical Analysis
The percentages of live insects were analyzed by SPSS statistical software v. 12.0 (SPSS Inc., Chicago, Illinois, United States). Data were subjected to a one-way analysis of variance (ANOVA), and comparisons between groups were performed with an analysis of Tukey's studentized range (t-test). Data were expressed as means ± standard errors (SEs), and statistical significance was set at the 0.05 (α) level of significance.
3 Results
3.1 Morphological Identification
3.1.1 Metarhizium robertsii KB6T1W1
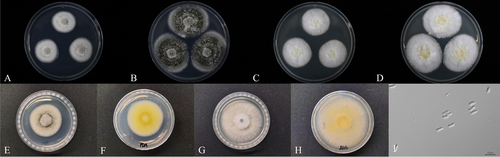
Colonies attained a diameter that ranged from 40 to 45 mm and 38–45 mm on PDA and SDA, respectively, within 14 days at 25°C. After Day 7, the colonies consisted of creamy white mycelium (Figure 1A). After 14 days on PDA, some powdery areas of dark greenish conidia gradually expanded and covered the entire area, and the edges showed a pale green color (Figure 1B), and the reverse side showed a light yellow color (Figure 1F). After 14 days on SDA, greenish conidia were not developed, and light white mycelium was still maintained (Figure 1C). The conidia were slightly clavate, smooth-walled, cylindrical with rounded apices, hyaline (Figure 1I), and measured 5.5–6.4 × 2.0–2.4 μm in size.
3.1.2 Metarhizium pinghaense IPBL-I
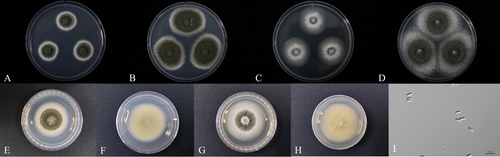
Colonies attained a diameter that ranged from 47 to 54 mm and 55–60 mm on PDA and SDA, respectively, within 14 days at 25°C. After 7 days, the colonies consisted of covered conidia as quickly grown dark greenish conidia, including white mycelium (Figure 2A). Until 14 days on PDA, dark greenish conidia changed olive color (Figure 2B), and the reverse side showed a light white color (Figure 2F). After 7 days on SDA, dark greenish conidia were in the center position, and the mycelium color was light white (Figure 2C). After 14 days olive color conidia expanded (Figure 2D). The conidia were cylindrical, slightly raised at the apex, smooth-walled (Figure 2I), and measured 5.5–6.4 × 2.0–2.4 μm in size.
3.1.3 Metarhizium lepidiotae KW9R2W1
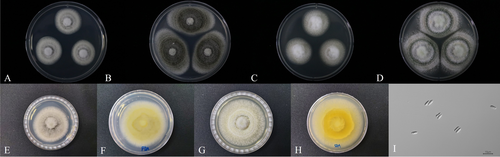
Colonies attained a diameter that ranged from 47 to 54 mm and 55–60 mm on PDA and SDA, respectively, within 10 days at 25°C. After 7 days, the colonies consisted of cotton-like white mycelium and some dark greenish conidia powdery on mycelium (Figure 3A). After 14 days, the dark greenish color changed to light olive and then to dark gray (Figure 3B), with the reverse side showing a slightly pale yellow color (Figure 3F). After 7 days on SDA, the colonies consisted of cotton-like white mycelium (Figure 3C). After 14 days, the conidia were greenish, the mycelium light gray (Figure 3D), and the reverse side showed a light yellow color (Figure 3H). Conidia were cylindrical with rounded apices, smooth-walled (Figure 3I), and 4.8–7.0 × 1.5–2.5 μm.
3.1.4 Metarhizium pemphigi IPBL-H
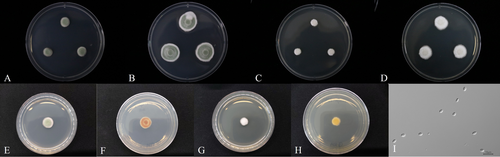
Colonies attained a diameter that ranged from 36 to 40 mm and 38–43 mm on PDA and SDA, respectively, within 14 days at 25°C. After 7 days, the colonies consisted of cotton-like white mycelium and some yellow-greenish conidia (Figure 4A). After 14 days, the yellow-green color appeared (Figure 4B), and the reverse side showed a slight pale yellow color (Figure 4F). After 7 days on SDA, the colonies consisted of cotton-like white mycelium (Figure 4C). After 14 days, conidia had not developed (Figure 4D), and the reverse side showed a whitish color (Figure 4H). Conidia were cylindrical, slightly raised at the apex, smooth-walled (Figure 4I), and measured 4.8–7.0 × 1.5–2.5 μm.

3.1.5 Metarhizium rileyi IPBL-J
Colonies attained a diameter that ranged from 36 to 40 mm and 38–43 mm on PDA and SDA, respectively, within 14 days at 25°C. After 7 days, the colonies consisted of velvet-shaped mycelium and light greenish conidia (Figure 5A). After 14 days, the colonies consisted of light greenish conidia with white mycelium at the edges (Figure 5B), and the reverse side showed an orange color (Figure 5F). After 7 days on SDA, the colonies consisted of cotton-like white mycelium (Figure 5C). After 14 days, conidia had not developed (Figure 5D), and the reverse side showed a light yellow color (Figure 5H). Conidia were broadly ellipsoidal, smooth-walled (Figure 5I), and measured 4.8–7.0 × 1.5–2.5 μm.
3.2 Identification and Phylogenetic Analyses
Morphological observation strongly inferred the six isolates (KW9R2W1, PBL-H, IPBL-I, IPBL-J, KB6T1W1) to the genus Metarhizium based on the formation of oval green conidia on the media (data not shown), and molecular identification was performed for more accurate identification.
For molecular phylogenetic analysis, TEF, ITS, and RPB1 sequences were used in this study (Table 3). The TEF dataset (877 bp) and combined dataset of concatenated multilocus sequences totaling 1843 bp (TEF 877 bp, RPB1 525 bp, and ITS 441 bp) were used in the analyses. As a result, a total of five species of Metarhizium were confirmed in this study (Figure 6); M. pinghaense IPBL-I, M. pemphigi IPBL-H, and M. rileyi IPBL-J were already discovered and recorded species, and M. robertsii KB6T1W1 and M. lepidiotae KW9R2W1were newly recorded species in Korea.
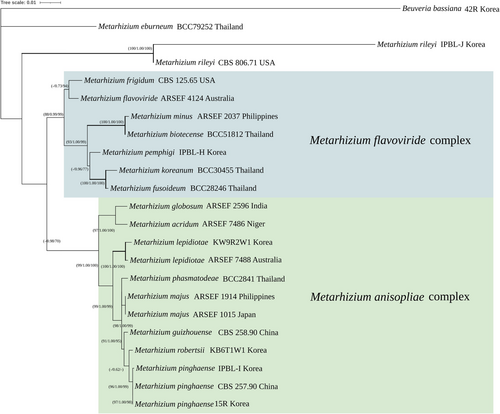
The concatenated sequences of three genes (TEF, RPB1, and ITS) were analyzed using MP, BI, and ML methods (Figure 1). BI (PP > 0.96) and ML analyses (MPBS > 96) strongly supported two clades of M. flavoviride complex and M. anisopliae complex. IPBL-I strains were in the clade as M. pinghaense. M. lepidiotae KW9R2W1 strain, which was not previously identified in Korea and made clade with M. lepidiotae ARSEF 7488, showed strong support value for three analysis methods. M. robertsii KB6T1W1 strain was confirmed as a relatively close sister group of M. pinghaense. M. pemphigi IPBL-H strain was located in M. flavoviride complex clade. M. rileyi IPBL-J strain made clade with M. rileyi CBS 806.71 strain.
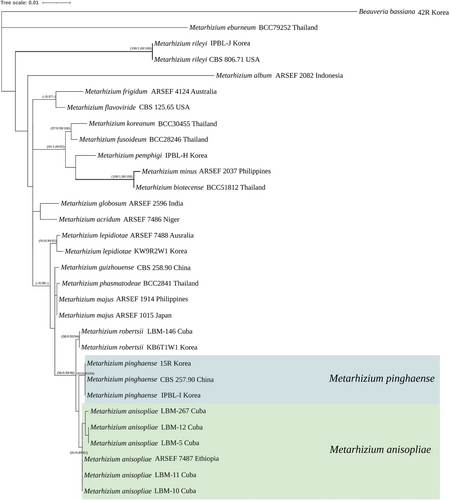
The results of single-gene TEF (877 bp) were analyzed using three methods (MP, BI, and ML) (Figure 7). The M. flavoviride complex and the M. anisopliae complex formed unclear and unresolved phylogenetic relationship between them. However, the TEF single gene tree is considered helpful in species identification. The 15R and IPBL-I strains are shown relatively close to M. pinghaense CBS 257.90.
3.3 Pathogenicity Bioassays Against Cotton Aphids and Lesser Mealworms
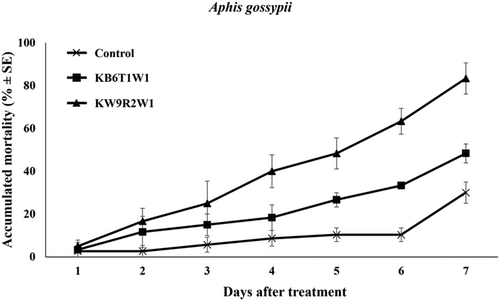
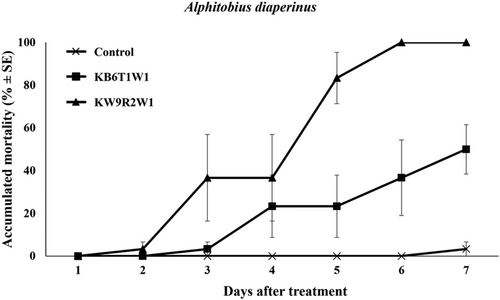
The pathogenicity of two unrecorded species in Korea, M. lepidiotae and M. robertsii, was evaluated against insect pests. M. lepidiotae KW9R2W1 demonstrated significantly higher pathogenicity compared to M. robertsii KB6T1W1 in both insect bioassays. In the cotton aphid (A. gossypii) bioassay, strain KW9R2W1 exhibited an 85% insecticidal effect (Figure 8), while strain KB6T1W1 resulted in a 55% insecticidal effect (F2, 42 = 56.82, p < 0.001). In the lesser mealworms (A. diaperinus) bioassay, M. lepidiotae KW9R2W1 achieved 100% mortality (Figure 9), whereas M. robertsii KB6T1W1 exhibited a 50% insecticidal effect (F2, 42 = 52.20, p < 0.001).
4 Discussion
Entomopathogenic fungi Metarhizium species and strain identification are important for biological control of insect pests. However, it is difficult to identify these species based on the morphological characteristics. Molecular identification is important in biotechnology and agriculture systems and has been used to develop new strains and determine biological diversity (Bischoff, Rehner, and Humber 2006, 2009; Driver, Milner, and Trueman 2000; Huang et al. 2005; Kepler et al. 2014; Kortsinoglou et al. 2019). The ITS region has traditionally been used for the identification of Metarhizium species, although ITS as a barcode marker will not aid in the proper identification of the species (Mongkolsamrit et al. 2020). In this study, the results of the analysis including ITS also generated a similar lineage to that in previous studies, so it is still thought to be useful for phylogenetic analysis rather than barcoding for species identification.
Molecular phylogenetic analysis including ITS showed a clade containing M. anisopliae and M. flavoviride complexes while the single gene TEF was used for Metarhizium species identification. As a result, the TEF gene was useful for species identification of Metarhizium species especially M. pinghaense and M. anisopliae.
In our knowledge, the molecular biological analysis method of this study confirmed that two species M. robertsii and M. lepidiotae, which had not been reported to live in Korea, were found to live in Korea. More Metarhizium species are anticipated to inhabit Korea if the genus Metarhizium, which has previously been reported to be isolated, is reidentified.
In bioassays conducted during this study, the entomopathogenic fungus M. lepidiotae KW9R2W1 demonstrated significant pathogenicity against both cotton aphids and lesser mealworms. However, during the preliminary screening phase, this strain exhibited relatively low pathogenicity at lower concentrations against cotton aphids, particularly at 1 × 106 conidia/mL (data not shown). This result indicates that M. lepidiotae KW9R2W1 may require a higher fungal concentration, such as 1 × 108 conidia/mL or more, to exert effective control. Conversely, the bioassays for the M. robertsii KB6T1W1 revealed minimal pathogenicity against cotton aphids, showing no significant impact on aphid mortality.
This variation in pathogenicity between strains may be attributable to intraspecific diversity within M. robertsii, as differences in strain-specific virulence, host specificity, or environmental adaptability are well documented in entomopathogenic fungi (Bidochka et al. 2001; Meyling and Eilenberg 2007). Previous studies have suggested that isolates from different geographical regions or ecological niches may exhibit distinct host preferences, which could explain the observed disparity in pathogenicity toward cotton aphids in this study (Bidochka et al. 2001; Meyling and Eilenberg 2007). For example, M. robertsii isolates from other regions have been reported to infect a wide range of insect hosts, ranging from species with exophthalmic to endophthalmic features. This indicates that even within the same species, the host range can vary depending on various factors such as the environment in which it evolved or became isolated.
This study serves as foundational research into Metarhizium species native to Korea. Our findings reveal a greater diversity of Metarhizium species in Korea than previously speculated, which had largely been represented by M. anisopliae complex and M. flavoviride complex. Notably, we identified two previously unreported Metarhizium species, expanding the known biodiversity of this genus in Korea. These results suggest the potential for even greater species diversity through continued investigation. The data generated in this study can provide an important baseline for future research on native Metarhizium species in Korea, with potential implications for biological control.
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agricultural Machinery/Equipment Localization Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321054-05).




