Rimonabant suppresses RNA transcription of hepatitis B virus by inhibiting hepatocyte nuclear factor 4α
Asuka Satoa and Chikako Ono contributed equally to this work.
Abstract
Chronic infection with hepatitis B virus (HBV) sometime induces lethal cirrhosis and hepatocellular carcinoma. Although nucleot(s)ide analogs are used as main treatment for HBV infection, the emergence of the drug-resistant viruses has become a problem. To discover novel antivirals with low side effects and low risk of emergence of resistant viruses, screening for anti-HBV compounds was performed with compound libraries of inhibitors targeting G-protein-coupled receptors (GPCRs). HepG2-hNTCP C4 cells infected with HBV were treated with various GPCR inhibitors and harvested at 14 day postinfection for quantification of core protein in the first screening or relaxed circular DNA in the second screening. Finally, we identified a cannabinoid receptor 1 inhibitor, rimonabant, as a candidate showing anti-HBV effect. In HepG2-hNTCP C4 cells, treatment with rimonabant suppressed HBV propagation at the viral RNA transcription step but had no effect on entry or covalently closed circular DNA level. The values of half maximal inhibitory concentration, half maximal effective concentration, and selectivity index of rimonabant in primary human hepatocyte (PHH) are 2.77 μm, 40.4 μm, and 14.6, respectively. Transcriptome analysis of rimonabant-treated primary hepatocytes by RNA sequencing revealed that the transcriptional activity of hepatocyte nuclear factor 4α (HNF4α), which is known to stimulate viral RNA synthesis, was depressed. By treatment of PHH with rimonabant, the expression level of HNF4α protein and the production of the messenger RNAs (mRNAs) of downstream factors promoted by HNF4α were reduced while the amount of HNF4α mRNA was not altered. These results suggest that treatment with rimonabant suppresses HBV propagation through the inhibition of HNF4α activity.
Abbreviations
-
- ALDOB
-
- aldolase B
-
- cccDNA
-
- covalently closed circular DNA
-
- CNR1
-
- cannabinoid receptor 1
-
- ETV
-
- entecavir
-
- FABP1
-
- fatty acid-binding protein 1
-
- GEq
-
- genome equivalents
-
- GPCR
-
- G-protein-coupled receptor
-
- HBc
-
- HBV core protein
-
- HBV
-
- hepatitis B virus
-
- HNF4α
-
- hepatocyte nuclear factor 4α
-
- NAs
-
- nucleot(s)ide analogs
-
- NTCP
-
- sodium taurocholate cotransporting polypeptide
-
- pgRNA
-
- pregenomic RNA
-
- PHH
-
- primary human hepatocyte
-
- rcDNA
-
- relaxed circular DNA
-
- SI
-
- selectivity index
1 INTRODUCTION
Hepatitis B virus (HBV) has chronically infected more than 250 million people worldwide.1 Chronic infection with HBV is a causative agent of cirrhosis and hepatocellular carcinoma, which together leads to more than 600,000 deaths per year.2 HBV, a member of the Hepadnaviridae family, possesses a partially double-stranded circular DNA genome. HBV particles are composed of an icosahedral nucleocapsid and envelope glycoproteins consisting of large, middle, and small S proteins. The preS1 region in the large S protein is involved in the entry into host cells through interaction with a receptor, sodium taurocholate cotransporting polypeptide (NTCP).3 Upon fusion of the HBV envelope with the host plasma membrane, the nucleocapsid is released into the host cytosol, then transported to the nuclear pores,4 and a complete double-stranded circular DNA described as covalently closed circular DNA (cccDNA) is generated after release of the viral genome into the nucleus. The cccDNA acts as a template for transcription of all viral RNAs, and several host factors are reported to be involved in this transcriptional step. Exogenous expression of hepatocyte nuclear factor 4α (HNF4α) and retinoid X receptor α/peroxisome proliferation activated receptor α in nonhepatic cell lines incapable of replicating HBV supports the RNA synthesis of HBV.5
Chronic infection with HBV is currently treated with nucleot(s)ide analogs (NAs), such as tenofovir and entecavir (ETV). NAs inhibit reverse transcription of pregenomic RNA (pgRNA) to relaxed circular DNA (rcDNA) through incorporation into the viral genome followed by DNA chain termination. However, the treatment has the potential to reactivate HBV upon discontinuation of therapy, because when viral cccDNA is no longer directly targeted by NAs, it can persist stably in host cells. In addition, the emergence of drug-resistant viruses has been reported following treatment with NAs.6 Therefore, novel strategies are needed for the treatment of chronic hepatitis B. To identify novel antivirals for HBV with a low risk of emergence of drug-resistant breakthrough viruses, we screened a compound library of G-protein-coupled receptor (GPCR)-associated drugs targeting host factors. GPCRs form one of the largest protein superfamilies and are known to transport the signals of nerve transmitter substances or hormones into cells.7 Previous studies have shown that GPCRs are involved in various viral life cycles and immune responses upon viral infection. Influenza A virus enters the target cells by endocytosis via free fatty acid receptor 2 signaling,8 and TGR5, a metabolite-sensing GPCR, is induced by viral infection and stimulates Type I IFN production via the AKT/IRF3 signaling cascade.9 Therefore, we expected that some GPCRs participate in the HBV life cycle or the immune system against HBV.
In this study, we found that rimonabant, a cannabinoid receptor 1 (CNR1) inhibitor, has an anti-HBV activity. Rimonabant inhibits RNA synthesis of HBV independent of CNR1 expression in HepG2-hNTCP C4 cells, Huh7 cells, and primary human hepatocytes (PHHs). Low expression of the HNF4α protein was observed in PHH treated with rimonabant, whereas expression of messenger RNA (mRNA) was not altered. Taken together, these results suggest that rimonabant suppresses HBV propagation by reduction of pgRNA synthesis through degradation of the HNF4α protein.
2 MATERIALS AND METHODS
2.1 Cell lines and viruses
All three hepatoma cell lines, HepG2-hNTCP C4 cells, Huh7 cells, and HepAD38.7 cells, and the PHH (Phoenix Bio, Hiroshima, Japan) were cultured at 37°C under the conditions of a humidified atmosphere and 5% CO2. As described in the previous report, Huh7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Nacalai Tesque, Kyoto, Japan) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (Sigma, St. Louis, MO), and 10% FBS; HepG2-hNTCP-C4 cells were maintained in the aforementioned medium containing 400 μg/ml G418 (Nacalai Tesque); and HepAD38.7 cells were cultured in DMEM/F-12 medium supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 18 μg/ml hydrocortisone (Sigma), 5 μg/ml insulin (Sigma), 400 μg/ml G418, and 400 ng/ml tetracycline (Nacalai Tesque). HBV was obtained from the culture supernatants of HepG2 transfected with plasmids containing a 1.3-fold-overlength genome of HBV genotype A, pUC19-Ae_US, or from the culture supernatants of HepAD38.7 cells that produce HBV when incubated in tetracycline-free medium. For HBV infection, HepG2-hNTCP-C4 cells were incubated overnight on 12 well plates (Iwaki, Tokyo, Japan) coated with collagen Type 1, and were inoculated with 10,000 genome equivalents (GEq)/cell of HBV in the aforementioned medium containing 3% DMSO (Sigma) and 4% PEG 8000 (Nacalai Tesque). The culture medium was replaced every 2 or 3 days.10 HepG2-hNTCP-C4 cells and HepAD38.7 cells were kindly provided by Dr T. Wakita, and the plasmid, pUC19-Ae_US, was kindly provided by Dr M. Mizokami and has been described previously.11 PHH was maintained in PHH-specific medium (Phoenix Bio). HBV genotype A, C (Phoenix Bio), or D infection into PHH was performed at 5 GEq per cell in the PHH-specific medium containing 4% PEG 8000 (Nacalai Tesque).
2.2 Immunoblotting
Cells were lysed on ice with lysis buffer containing 20 mm Tris-HCl (pH 7.4), 135 mm NaCl, 1% Triton-X 100, and 10% glycerol, boiled in loading buffer for 10 min, and then resolved by 5%–20% gradient SDS-PAGE. The proteins were transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA), blocked with PBS consisting of 0.05% Tween 20 and 5% skim milk, and reacted with the appropriate antibodies at room temperature. The immune complexes were visualized with Super Signal West Femto Substrate (Pierce, Rockford, IL) and detected using an Amersham Imager 600UV image analyzer system (Fujifilm, Tokyo, Japan).
2.3 Antibodies and compounds
Mouse anti-HBc antibody was kindly provided by Dr A. Ryo. Anti-CNR1 (C-Term) rabbit polyclonal antibody (Cayman, Ann Arbor, MI), HRP-conjugated rabbit monoclonal antibody, mouse monoclonal antibody to β-actin, and HRP-conjugated mouse monoclonal antibody were purchased from Sigma, and anti-HNF4α (C11F12) rabbit monoclonal was purchased from Cell Signaling. The GPCR compound library (96 well) used for screening was purchased from TargetMol (Shanghai, China), and rimonabant and AM251 were purchased from Selleckchem (Houston, TX).
2.4 Purification of intracellular HBV rcDNA and cellular RNA
As we showed in our previous report, intracellular HBV DNA was extracted as follows: First, the cell pellets were lysed by lysis buffer (50 mm Tris-HCl [pH 7.4], 1 mm EDTA, 1% NP-40) at 4°C for 15 min. After centrifugation at 15,000 rpm for 5 min, the supernatant was incubated with 7 mm magnesium acetate, 0.2 mg/ml of Dnase I (Roche, Mannheim, Germany), and 0.1 mg/ml of RNase A (Sigma) at 37°C for 3 hr. After the addition of 10 mm EDTA and 10 mm NaCl, the lysates were digested by proteinase K (0.3 mg/ml; Thermo Fisher Scientific, Waltham, MA) and 2% SDS at 55°C for 5 hr. The extracted HBV DNA was purified by phenol–chloroform–isoamyl alcohol, precipitated with ethanol, and resolved in pure water.10 Total RNA was extracted by using a Pure Link RNA Mini Kit (Thermo Fisher Scientific) according to the manufacturer's protocol.
2.5 Quantitative polymerase chain reaction
qPCR was performed for HBV rcDNAs and pgRNAs using Fast SYBR green master mix (Applied Biosystems, Foster City, CA) with the primer pairs 5′-GGAGGGATACATAGAGGTTCCTTGA-3′ (forward) and 5′-GTTGCCCGTTTGTCCTCTAATTC-3′ (reverse), and 5′-TCCCTCGCCTCGCAGACG-3′ (forward) and 5′-GTTTCCCACCTTATGAGTC-3′ (reverse),12 respectively; for HBV cccDNAs using TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) with the primer pair 5′-CGTCTGTGCCTTCTCATCTGC-3′ (forward) and 5′-GCACAGCTTGGAGGCTTGAA-3′ (reverse), and a probe 5′-CTGTAGGCATAAATTGGT-3′; and for cellular RNAs using a TaqMan RNA-to-Ct one-step kit and ViiA7 real-time PCR system (Thermo Fisher Scientific). The following cellular RNAs were detected by TaqMan Gene Expression Assay (FAM): HNF4α (Assay ID: Hs00604435_m1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Hs02758991_g1), aldolase B (ALDOB; Hs01551887_m1), and fatty acid-binding protein 1 (FABP1; Hs00155026_m1). The expression level of pgRNA and each cellular gene was determined by the ΔΔCT (where CT is the threshold cycle) method using GAPDH as an internal control.13
2.6 Interaction assay of hNTCP with preS1
For flow cytometry experiments, HepG2-hNTCP-C4 cells (2.5 × 105 cells) were suspended in PBS containing 2% FBS and incubated on ice with compounds at 10 μm for 30 min. Fluorescence-labeled PreS1 peptide solution was added to the cells, and the cells were incubated on ice for 30 min. The cell solutions were then supplemented with 3 ml of 2% FBS/PBS and centrifuged at 1,500 rpm for 3 min, and the supernatants were aspirated. The samples were analyzed by a FACSCalibur flow cytometer (Nippon Becton Dickinson, Tokyo, Japan) and FlowJo 7.6.5 software.
2.7 Transfection
The plasmids containing a 1.3-fold-overlength genome of HBV genotype B, pGEM-Bj-JPN56 (accession no. AB246342), or genotype C, pUC19-C_JPNAT (accession no. AB246345), were kindly provided by Dr M. Mizokami and have been described previously.11 These plasmids were transfected into Huh7 cells using Trans IT LT-1 transfection reagent (Mirus, Madison, WI).
2.8 Cell viability assay
The HepG2-hNTCP-C4 cells were seeded on 96 well plates (Iwaki) coated with collagen Type 1 and incubated overnight. Viabilities of the cells were measured by a luminescent cell viability assay at 14 days postinfection with exposure to the compounds.
2.9 RNA sequencing
The data were acquired following the procedure in a previous report, as described below. Total RNA in PHHs was extracted using a PureLink RNA Mini Kit (Thermo Fisher Scientific). RNA libraries were prepared for sequencing using a TruSeq Stranded Total RNA with Ribo-Zero kit (Illumina, San Diego, CA). Whole transcriptome sequencing was applied to RNA samples using the Illumina HiSeq 2500 platform in a 75 bp single-end mode. Sequenced reads were mapped to the human reference genome sequence (hg19) using TopHat version 2.0.13 in combination with Bowtie 2 version 2.2.3 and SAMtools version 1.0. The number of fragments per kilobase of exon per million mapped fragments was calculated using Cuffnorm version 2.2.1.14 The raw data from this study were submitted to the Gene Expression Omnibus and are available under accession number GSE139597.
2.10 Statistics
The results are exhibited as the means ± standard deviation. The significance of differences in means was determined by Student's t-test.
3 RESULTS
3.1 Effect of GPCR-associated drugs on HBV replication
To identify a novel compound showing an anti-HBV effect, we screened a library of 533 GPCR compounds. As shown in Figure 1a, HepG2-hNTCP C4 cells stably expressing NTCP on the surface and susceptible to HBV infection were treated with each compound at 10 μm from 6 hr before to 1 day postinfection with HBV, and 7–10 days postinfection to identify compounds inhibiting one of the steps from entry to replication of the viral genome in the HBV life cycle. At 14 days postinfection, quantitative Western blotting of the intracellular HBV core protein (HBc) was carried out for the samples showing a high cell-survival rate, which was determined by counting surviving cells. In the first screening, 56 of 533 compounds suppressed the expression of HBc to less than 30% of that in the control treated with DMSO (Figure 1b, left). In the second screening, intracellular HBV rcDNA was measured by qPCR, and 19 of 56 compounds inhibited rcDNA expression to less than 60% of that in the negative control (Figure 1b, right). The 19 compounds included two CNR1 inhibitors, rimonabant and AM251, and we therefore decided to focus on CNR1 inhibitors in this study. The GPCR compound library we used includes another CNR1 inhibitor, AM281. AM281 has a chemical structure similar to those of rimonabant and AM251 (Figure 1c), and all three compounds exhibited anti-HBV effects (Figure 1d), suggesting that CNR1 inhibitors possess anti-HBV activity.
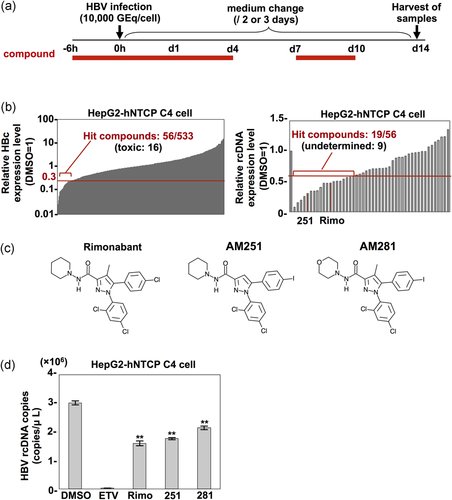
3.2 Rimonabant inhibits transcription of HBV RNA
To determine the mechanisms by which rimonabant inhibits HBV propagation, HepG2-hNTCP C4 cells treated with rimonabant at various time points were infected with HBV, and intracellular HBV rcDNA was quantified at 14 days postinfection (Figure 2a). Rimonabant significantly inhibited the rcDNA level even when administered at 1 day postinfection (Figure 2b, Condition III). In addition, rimonabant exhibited no effect on the binding between the receptor and preS1, which is located in the amino-terminal region of the large HBV surface protein and involved in the cell attachment and entry (Figure 2c), suggesting that rimonabant does not participate in viral entry. Next, to examine the effect of rimonabant on transcription and replication of the viral genome, cccDNA, rcDNA, and pgRNA production in HepG2-hNTCP C4 cells infected with HBV was quantified by qPCR. As shown in Figure 2d,e, treatment with heparin, which inhibits HBV attachment to the cells, suppressed cccDNA synthesis and pgRNA transcription. By contrast, production of rcDNA but not cccDNA or pgRNA was reduced in the cells treated with ETV, which inhibits reverse transcription from pgRNA to rcDNA, indicating the validity of this cccDNA and pgRNA measurement system. Rimonabant exhibited no effect on cccDNA synthesis but reduced pgRNA expression (Figure 2d,e), suggesting that it suppresses HBV propagation by inhibiting pgRNA transcription. To further confirm the inhibitory effect of rimonabant on other genotypes of HBV, Huh7 cells were transfected with 1.3× HBV genome plasmids of genotypes B and C, and the intracellular rcDNAs were examined. As expected, the expressions of intracellular rcDNA of both genotypes also significantly declined (Figure 2f). Taken together, these results suggest that rimonabant suppresses HBV propagation via inhibition of RNA transcription.
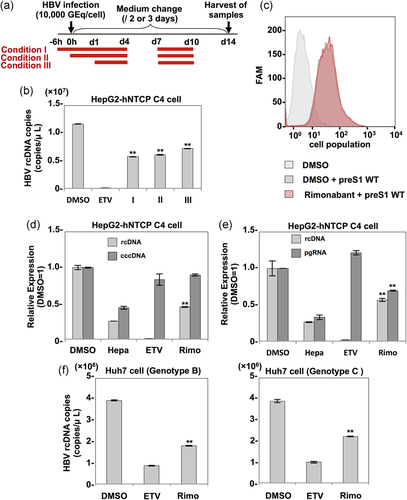
3.3 Anti-HBV activity of rimonabant in HepG2-hNTCP C4 cells and PHHs
Next, to examine the anti-HBV effect of rimonabant in HepG2-hNTCP C4 cells and PHH, these cells were treated with several concentrations of rimonabant from 6 hr before infection with HBV. The intracellular rcDNA and cell viability of both cell types at 14 days postinfection were determined by qPCR and CellTiter-Glo, respectively. The half maximal effective concentration (EC50), 50% cytotoxic concentration, and selectivity index (SI) values for rimonabant were 9.92 μm, 47.8 μm, and 4.82 in HepG2-hNTCP C4 cells, and 2.77 μm, 40.4 μm, and 14.6 in PHH, respectively (Figure 3a). The notable difference in EC50 values between HepG2-hNTCP C4 cells and PHH might be attributable to the difference in some metabolic systems between hepatoma cell lines and primary hepatocytes. When PHHs were treated with rimonabant from 6 hr before infection (Condition I), concurrent with infection (Condition II), or 1 day postinfection (Condition III; Figure 3b), even treatment at 1 day postinfection (Condition III) suppressed HBV production by approximately 75% (Figure 3c). When we exposed PHH to DMSO, heparin, ETV, or rimonabant from 6 hr before infection to 14 days postinfection, cccDNA synthesis in heparin-, ETV-, or rimonabant-treated cells was remarkably suppressed compared with that in DMSO-treated cells (Figure 3d). However, the production level of cccDNA in rimonabant-treated cells was significantly higher than that in the cells treated with heparin, which can suppress cccDNA synthesis before it is initiated by inhibiting the HBV entry step, and comparable to that in the cells exposed to ETV, which indirectly suppresses cccDNA synthesis by inhibiting rcDNA recycling after stable cccDNA persistence is established in PHH (Figure 3d). These results suggest that rimonabant inhibits HBV propagation in PHH via its effects on steps other than entry and cccDNA synthesis. To further investigate the anti-HBV effect of rimonabant on other genotypes of HBV, PHHs treated with rimonabant were infected with HBV of genotypes A and C. Rimonabant suppressed the propagation of HBV of both genotypes (Figure 3e). In addition, we examined the anti-HBV effect of the other CNR1 inhibitors in PHHs, and we found that both AM251 and AM281 suppressed HBV propagation to a degree comparable to rimonabant (Figure 3f). Finally, we examined the inhibitory effect of rimonabant against the late stage of HBV infection. HepG2-hNTCP C4 cells were exposed to 0.3, 1, 3, 10, or 30 μm rimonabant at 7–10 days postinfection (Figure 3g, upper left panel), and PHH was treated with 10 μm rimonabant at 7–14 days postinfection (Figure 3g, upper right panel). In HepG2-hNTCP C4 cells, treatment with 10 μm rimonabant reduced rcDNA production by approximately 20% compared with the control level and treatment with 30 μm rimonabant reduced rcDNA production to a level equivalent to that by treatment with 10 μm rimonabant from 6 hr before infection (Figure 3g, lower left panel). By contrast, treatment of PHH from 7 days postinfection suppressed HBV propagation by approximately 60% (Figure 3g, lower right panel). These data suggest that rimonabant possesses an anti-HBV effect against the late stage of HBV infection.
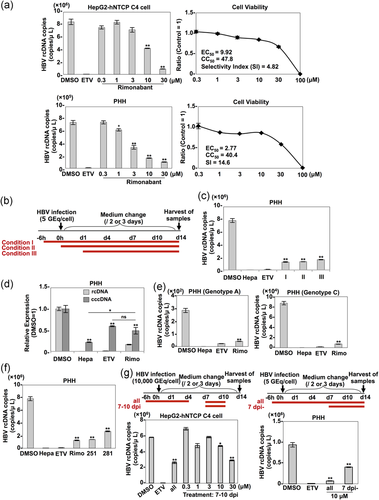
3.4 Treatment of PHH with rimonabant downregulated the transactivation of HNF4α required for HBV pgRNA transcription
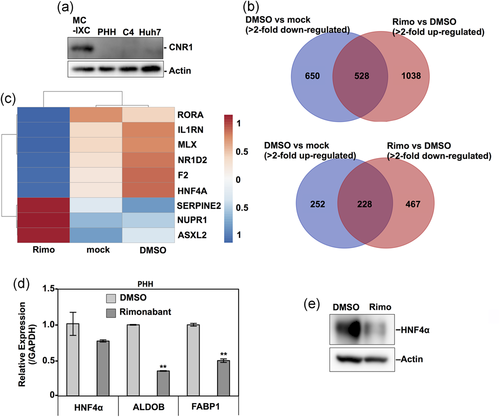
A previous study reported that CNR1, a target of rimonabant, is expressed at low levels in the normal human liver,15 but we could not detect any expression of CNR1 in PHH, HepG2-hNTCP C4, or Huh7 cells (Figure 4a), suggesting that rimonabant suppresses HBV propagation independent of CNR1 inhibition. To examine the underlying mechanisms by which rimonabant inhibits HBV propagation, transcriptome analysis was performed using an RNA sequence of nontreated (mock) PHH or PHH treated with either rimonabant or DMSO. The number of genes which showed an altered expression in DMSO-treated cells that was cancelled by treatment with rimonabant was 756 (upregulated or downregulated genes were 528 or 228, respectively; Figure 4b). Based on this change of gene expression in the gene data sets, an upstream regulator analysis was performed to predict the activation or inhibition of transcription factors and the direction of changes in expression by treatment with rimonabant using Ingenuity Pathway Analysis software (QIAGEN, Redwood City, CA; www.qiagen.com). The prediction was established based not on changes in the expression of the upstream regulator itself but rather on changes in the z-score from the activation z-score algorithm. This approach is quite different from computational promoter analysis of over-represented cis-motifs residing within the 5′-promoter regions of this gene set.16 This analysis predicted some significantly activated or inhibited pathways, as shown in Figure 4c. Because these pathways did not include IFN-stimulated response elements involved in the IFN signaling (Figure 4c), rimonabant appeared to suppress the HBV propagation independently of the IFN signaling pathway. By contrast, the HNF4α-associated pathway was significantly downregulated in PHH treated with rimonabant (Figure 4c). HNF4α has been shown to be an important transcriptional factor of HBV RNA,17 suggesting that HNF4α participates in the rimonabant-mediated anti-HBV effect. Although comparable expression of HNF4α mRNA was detected in PHH treated with rimonabant (Figure 4d), the HNF4α protein and mRNA expressions of two downstream factors of HNF4α—that is, ALDOB and FABP118—were significantly reduced by the treatment with rimonabant (Figure 4d,e). These results suggest that rimonabant regulates HNF4α protein expression without affecting the HNF4α mRNA expression, and thereby suppresses HBV RNA transcription.
4 DISCUSSION
NAs are currently the main therapeutics for chronic hepatitis B patients, but treatment with these drugs can induce the emergence of drug-resistant viruses. Thus, reducing the risk of inducing resistant viruses is one of the important goals in the development of anti-HBV drugs. In this study, we identified rimonabant as a candidate for a novel anti-HBV drug capable of inhibiting pgRNA synthesis of HBV by depressing the transcriptional activity of HNF4α.
HNF4α is known to promote pgRNA synthesis by interacting with the putative binding site of the HBV core promoter.19 Therefore, various anti-HBV compounds targeting HNF4α have been reported. For example, luteolin, a flavonoid with immune-regulatory and anti-inflammatory activities, and KX2-391, an Src inhibitor, exhibit anti-HBV effects by reducing the HNF4α mRNA level.19, 20
In this study, we showed that treatment of PHH with rimonabant suppressed HNF4α expression, but had no significant effect on the mRNA expression (Figure 4d,e). Expression of HNF4α is regulated by several different steps, including translation and proteolysis. The cooperative effects of G-quadruplex in and adjacent putative protein-binding sites within the 5′ untranslated region of HNF4α mRNA mediate translational repression of the nuclear receptor.21 Adenosine monophosphate-activated protein kinase regulates HNF4α activity by inhibiting the dimer formation of HNF4α and decreasing the stability of the protein.22 Ser78 phosphorylation of HNF4α by protein kinase C promotes degradation of HNF4α via a proteasome-dependent pathway.23 PolySUMOylated HNF4α enhances ubiquitination by RNF4, resulting in the ubiquitin-mediated degradation.24 One or more of these mechanisms might be involved in the depression of HNF4α by the treatment with rimonabant. Moreover, in this study, we only focused on HNF4α as a factor participating in the anti-HBV effect of rimonabant and did not examine the involvement of other factors for which significant activation or inhibition was predicted by transcriptome analysis because we could not find any studies reporting relevance between HBV and the other identified factors. Therefore, the possibility that the genes other than HNF4α affect the anti-HBV activity of rimonabant remains.
Rimonabant was originally discovered as a selective CNR1 blocker. The CNR1 protein exists in a high concentration in the brain, especially in the hypothalamic areas, which are known to control food intake and feeding behavior and to participate in the mesolimbic dopamine pathway, that is, the so-called reward system.25 Therefore, research into CNR1 has focused on this protein's direct association with feeding regulation and indirect involvement in the dopamine-mediated rewarding properties of food, leading to the development of CNR1 antagonists for the treatment of obesity. Rimonabant was first approved in Europe in 2006 as an orally active anorectic antiobesity drug; however, the approval was withdrawn in 2008 due to serious psychiatric side effects, and its development was terminated.26, 27 For future clinical use of rimonabant as an anti-HBV drug, several obstacles remain. The first is the psychiatric side effects that led to withdrawal of the marketing authorization of rimonabant. To overcome the obstacles, it might be useful to modify the tropism of rimonabant from the central nervous system to the liver, but even so, potential risks could remain. In fact, rimonabant treatment results in significant downregulation of ALDOB and FABP1 (Figure 4d), which participate in glycolysis and gluconeogenesis28 and in fatty acid uptake, transport, and metabolism in the liver,29 respectively. Therefore, further in vivo studies are needed to assess the appropriate dosages of rimonabant to suppress HBV replication but not liver function.
ACKNOWLEDGMENTS
The authors would like thank M. Tomiyama for her secretarial work and M. Ishibashi for her technical assistance. They are also grateful to T. Wakita, A. Ryo, and M. Mizokami for providing experimental materials. This work was supported by AMED under grant numbers JP19fk0310119, JP19fk0310102, JP19fk0310111, and JP19fk0310107.
DISCLOSURE
The authors declare no conflicts of interest associated with this manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.