Reassortant DS-1-like G1P[4] Rotavirus A strains generated from co-circulating strains in Vietnam, 2012/2013
ABSTRACT
One major mechanism by which Rotavirus A (RVA) evolves is genetic reassortment between strains with different genotype constellations. However, the parental strains of the reassortants generated have seldom been identified. Here, the whole genome of two suspected reassortants, RVA/Human-wt/VNM/SP127/2013/G1P[4] and RVA/Human-wt/VNM/SP193/2013/G1P[4], with short RNA electropherotypes were examined by Illumina MiSeq sequencing and their ancestral phylogenies reconstructed. Their genotype constellation, G1-P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2, indicated that they were G1 VP7 mono-reassortants possessing DS-1-like genetic backbones. The two strains were ≧99.7% identical across the genome. While their VP7 genes were ≧99.7 identical to that of a Wa-like strain RVA/Human-wt/VNM/SP110/2012/G1P[8] which co-circulated during the 2012/2013 season, 10 genes were ≧99.8% identical to that of the DS-1-like strains RVA/Human-wt/VNM/SP015/2012/G2P[4] (and SP108) that co-circulated during the season. The identities were consistent with the phylogenetic relationships observed between the genes of the reassortants and those of the afore-mentioned strains. Consequently, the G1P[4] strains appear to have been generated by genetic reassortment between SP110-like and SP015-like strains. In conclusion, this study provides robust molecular evidence for the first time that G1P[4] strains detected in Hanoi Vietnam were generated by inter-genogroup reassortment between co-circulating G1P[8] and G2P[4] strains within the same place and season.
List of Abbreviations
-
- MEGA
-
- Molecular Evolutionary Genetics Analysis
-
- NSP
-
- non-structural protein
-
- RVA
-
- Rotavirus A
-
- VP
-
- viral protein
Rotavirus A (RVA), a member of the genus Rotavirus within the family Reoviridae, is the leading cause of severe infantile gastroenteritis worldwide 1. The RVA genome consists of 11 segments of double-stranded RNA that encode six structural viral proteins (VP1–VP4, VP6 and VP7), and six non-structural proteins (NSP1–NSP5/6) 1. Rotaviruses are dually classified into P and G genotypes based on the two outermost capsid proteins VP4 and VP7, respectively. According to previous reports 2-4 and the latest update from the Rotavirus Classification Working Group 5 (June, 2017), at least 50 P-genotypes and 35 G genotypes have been identified in humans and animals. Five G/P type combinations in humans namely G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8] are globally common 6, 7.
The dual classification system of RVA strains has been extended to include the other nine genome segments; thus, the whole genome of RVA strains VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6 is denoted by the descriptor Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx where x represents the genotype number 2, 3, 8. Most human RVA strains can be classified into two major and one minor genotype constellations, the Wa-like, DS-1-like and AU-1-like genotype constellations, which are described as G1/3/4/9-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1, G2-P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2 and G3-P[9]-I3-R3-C3-M3-A3-N3-T3-E3-H3, respectively 2, 3, 8, 9.
Inter-genotype reassortment events occasionally occur between strains possessing the Wa-like genotype constellation and those possessing the DS-1-like genotype constellation, leading to unusual combinations of G and P genotypes such as G1P[4], G3P[4] and G2P[8] 10-12. Recently, double-gene reassortants such as G1P[8] strains possessing the DS-1-like inner capsid and non-structural protein genes emerged 13-16. These reassortant strains were found to have completely swapped the DS-1-like outer capsid genes with those of the Wa-like strains. Despite the fact that these reassortants were hypothesized to be generated from locally co-circulating strains, the parental strains from which the single-gene reassortants or double-gene reassortants were generated have rarely been identified as co-circulating strains 13-17.
In a recent molecular epidemiological study conducted in Hanoi, Vietnam between 2012 and 2013, we reported the emergence and spread of DS-1-like G1P[8] double-gene reassortant strains which were apparently of clonal origin. We showed that the backbone genes of the double-gene reassortant strains were highly identical to one locally co-circulating G2P[4] strain detected during the study period 16. In addition, we detected RVA strains possessing the G1P[4] G/P combination in five specimens, two of which possessed identical electropherotypes that were similar to the electropherotype of the G2P[4] strains circulating during the study period. The aim of this study was therefore to determine the whole genome sequences of the two G1P[4] RVA strains to determine whether they were generated as a result of a reassortment event between the G2P[4] and G1P[8] strains circulating at the same time and place of detection as the G1P[4] strains.
MATERIALS AND METHODS
Study specimens
SP127 and SP193, the two G1P[4] specimens characterized in this study, were among RVA-positive specimens collected from November 2012 to June 2013 from children hospitalized for diarrhea in Saint Paul Hospital and Bac Mai Hospital in Hanoi, Vietnam 16. Initial identification of RVA was performed by Nakagomi et al. (16) using ELISA (Premier Rotaclone; Meridian Bioscience, Cincinnati, OH, USA) and G/P typing.
A G1P[8] strain (SP110) and two G2P[4] strains (SP015 and SP108) that had previously been sequenced by the Sanger method 16 were included as parental strain candidates; their sequences are available under the GenBank accession numbers LC066169–LC066191. SP110 shares the same electropherotype with 33 (67%) of the 49 G1P[8] strains possessing long RNA patterns during the study period, whereas SP015 and SP108 share the same electropherotype with 44 (88%) of the 50 G2P[4] strains possessing short RNA patterns 16.
RNA extraction, electropherotyping, cDNA library preparation and Illumina MiSeq sequencing
Viral RNA was extracted from 10% stool suspensions (w/v) using TRIzol LS Reagent (Life Technologies, Grand Island, NY, USA) and a Direct-zol RNA MiniPrep Kit (ZYMO Research, Irvine, CA, USA) in accordance with the manufacturer's protocol. The electrophoretic migration pattern of the study strains was determined on polyacrylamide gel as previously described by Gauchan et al. 18. Briefly, RNA was electrophoresed on a 10% polyacrylamide gel (0.75 mm thick) with 4% stacking gel using Laemmli's discontinuous buffer system and SE600 Ruby electrophoresis apparatus (GE Healthcare Bio-Science, Piscataway, NJ, USA) for 16 hr at a constant current of 8 mA per gel. Gels were stained with 0.19% (w/v) AgNO3 and developed in 3% (w/v) NaOH containing 0.5% (v/v) formaldehyde. Genomic RNA from strains Wa (G1P[8]) and KUN (G2P[4]) were included as reference strains for long and short RNA patterns, respectively.
For cDNA library preparation, the concentration of RNA in each sample was quantified using a NanoDrop spectrophotometer ND–1000 (NanoDrop Technologies, Wilmington, DE, USA) according to the manufacturer's instructions. The starting RNA concentration was normalized to 100 ng for all samples. Following the manufacturer's instructions and methods described previously 19-21, a 200 bp fragment library ligated with bar-coded adapters was prepared for the G1P[4] strains using a NEBNext Ultra RNA library Prep Kit for Illumina v1.2 (New England Biolabs, Ipswich, MA, USA) and a NEBNext Multiplex Oligos for Illumina (New England Biolabs). The cDNA library was purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Pasadena, CA, USA) according to recommendations in the NEBNext protocol. The quantity and the quality of the purified libraries were determined on a Qubit 2.0 flourometer (Invitrogen, Carlsbad, CA, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany), respectively.
Nucleotide sequencing was performed on an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA) using a MiSeq Reagent Kit v3 (Illumina) to generate 301 paired-end reads.
A templated whole genome assembly workflow available in the SeqMan NGen DNAStar program (SeqMan NGen. Version 12.0. DNASTAR; Madison, WI, USA) was adopted for assembling contigs for the study strains. Briefly, sequence reads were assembled into contigs by aligning reads to the genomes of RVA/Human-wt/BGN/M292/2013/G2P[4] and RVA/Human-wt/VNM/SP108/2012/G2P[4] (GenBank accession numbers KU356640–KU356650, LC0661810–LC066190, respectively) using the reference-based assembly normal workflow option. The maximum total reads was set within the range of 30,000–150,000 taking into account the rotavirus genome total size of 18,550 bp. Further downstream analysis was carried out using the SeqMan Pro Version 14 (DNASTAR). The number of reads for each genome segment are provided in Supplementary Table S1.
Genotyping and phylogenetic analysis
The genotypes of the contigs for each genome segment were determined using an automated online genotyping tool for RVA RotaC 2.0 22. Similar sequences possessing the same genotype were retrieved from the GenBank using the Basic Local Alignment Search Tool 23 and the genes of strain SP127 as the query sequences. Multiple sequence alignments were constructed using the MUSCLE program in MEGA v.6.06 24.
To investigate the possible parental strains of the G1P[4] reassortant strains, phylogenetic trees were constructed using datasets that included sequences of strains circulating in Vietnam and elsewhere in the world. It was hypothesized that the G1P[4] strains had been generated through a reassortment event between a G1P[8] and a G2P[4] strain in which the G2P[4] strain had acquired the G1 VP7 gene from a co-circulating G1P[8] strain. In this regard, the origin of the VP7 and VP4 genes was investigated first, after which the G2P[4] strain that had donated the remaining nine genes was searched for by employing phylogenetic analysis of the concatenated open reading frames of the VP6, VP1-VP3 and NSP1–NSP5 genes. Genetic distances were calculated by the p-distance method implemented in MEGA v. 6.06 24.
Nucleotide sequence accession numbers
The nucleotide sequences were deposited in the DDBJ/EMBL/GenBank database under the accession numbers LC215251–LC215272 and are listed in Table 1.
| RVA/Human-wt/VNM/SP127/2013/G1P[4] | RVA/Human-wt/VNM/SP193/2013/G1P[4] | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Genotype | Region of gene sequenced | % sequenced | Nucleotide sequence Accession number | Region of gene sequenced | % sequenced | Nucleotide sequence Accession number |
| VP7 | G1 | 1 to 1062 | 100 | LC215251 | 1 to 1062 | 100 | LC215262 |
| VP4 | P[4] | 1 to 2359 | 100 | LC215252 | 1 to 2359 | 100 | LC215263 |
| VP6 | I2 | 1 to 1356 | 100 | LC215253 | 1 to 1356 | 100 | LC215264 |
| VP1 | R2 | 1 to 3302 | 100 | LC215254 | 1 to 3302 | 100 | LC215265 |
| VP2 | C2 | 11 to 2684 | 99.6 | LC215255 | 10 to 2684 | 99.6 | LC215266 |
| VP3 | M2 | 1 to 2592 | 100 | LC215256 | 1 to 2591 | 100 | LC215267 |
| NSP1 | A2 | 1 to 1566 | 100 | LC215257 | 1 to 1566 | 100 | LC215268 |
| NSP2 | N2 | 11 to 1059 | 99.1 | LC215258 | 10 to 1059 | 99.2 | LC215269 |
| NSP3 | T2 | 1 to 1066 | 100 | LC215259 | 1 to 1066 | 100 | LC215270 |
| NSP4 | E2 | 1 to 751 | 100 | LC215260 | 1 to 751 | 100 | LC215271 |
| NSP5 | H2 | 9 to 816 | 99 | LC215261 | 1 to 816 | 100 | LC215272 |
- A, interferon antagonist; C, core shell; E, enterotoxin; H, pHosphoprotein; I, intermediate capsid shell; M, RNA-capping methyltransferase; N, octameric NTPase; R, RNA polymerase; T, Translation regulation.
RESULTS
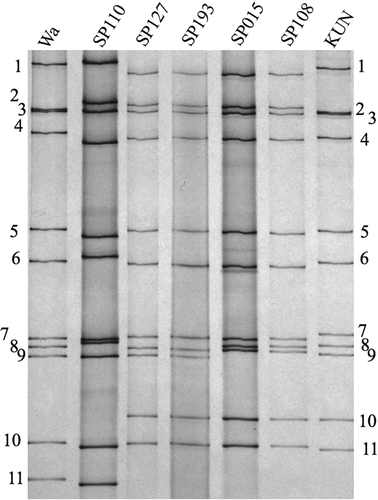
The study strains SP127 and SP193 were found to possess identical short RNA electropherotypes by polyacrylamide gel electrophoresis (Fig. 1). The complete or near complete lengths (99.0–100%) of the 11 genes of SP127 and SP193 were obtained (Table 1). Both strains possess the same genotype constellation G1-P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2. Their genotype constellation indicates that both strains were generated by genetic reassortment between G1P[8] and DS-1-like G2P[4] strains. Nucleotide sequence comparison between the 11 genes of SP127 and SP193 revealed high identities ranging from 99.7–100% (Table 2); indicating that they are the same strain.
| RVA/Human-wt/VNM/SP127/2013/G1P[4] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | ||
| Strain name | G1 | P[4] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 | Reference |
| SP193 | 99.7 | 99.7 | 99.9 | 99.9 | 99.8 | 99.8 | 99.9 | 99.9 | 99.9 | 99.7 | 100 | This study |
| SP108 | G2 | 99.9 | 99.9 | 99.9 | 99.9 | 99.8 | 99.9 | 99.9 | 99.8 | 99.9 | 100 | (15) |
| SP015 | G2 | 99.9 | 99.8 | 99.9 | 99.9 | 99.9 | 99.9 | 100 | 100 | 99.9 | 100 | (15) |
| SP110 | 99.8 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 | (15) |
| RVA/Human-wt/VNM/SP193/2013/G1P[4] | ||||||||||||
| VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | ||
| Strain name | G1 | P[4] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 | Reference |
| SP127 | 99.7 | 99.7 | 99.9 | 99.9 | 99.8 | 99.8 | 99.9 | 99.9 | 99.9 | 99.7 | 100 | This study |
| SP108 | G2 | 99.9 | 100 | 99.9 | 99.8 | 99.9 | 99.9 | 100 | 99.9 | 99.9 | 100 | (15) |
| SP015 | G2 | 99.8 | 99.8 | 99.8 | 99.8 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 100 | (15) |
| SP110 | 99.7 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 | (15) |
- A, interferon antagonist; C, core shell; E, enterotoxin; H, pHosphoprotein; I, intermediate capsid shell; M, RNA-capping methyltransferase; N, octameric NTPase; R, RNA polymerase; T, Translation regulation.
Regarding the parental strains of SP127 and SP193, it was found that their G1 VP7 gene was respectively 99.8% and 99.7% identical to SP110, a G1P[8] strain that had circulated during the season (16) (Table 2). The remaining ten genes of SP127 and SP193 were ≧99.8% identical to the genes of the G2P[4] strains SP015 and SP108 that co-circulated during the study period (16) (Table 2); this observation was consistent with their identical electrophoretic migration patterns (Fig. 1).
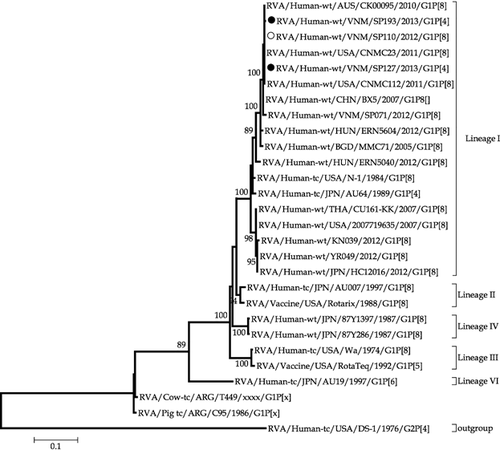
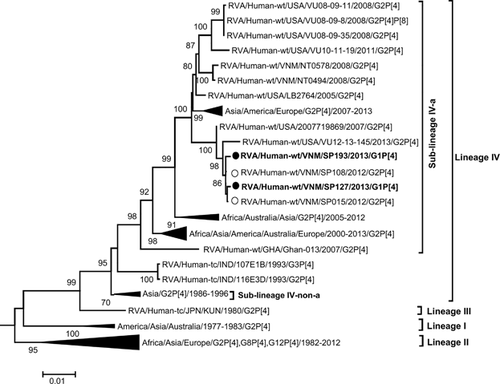
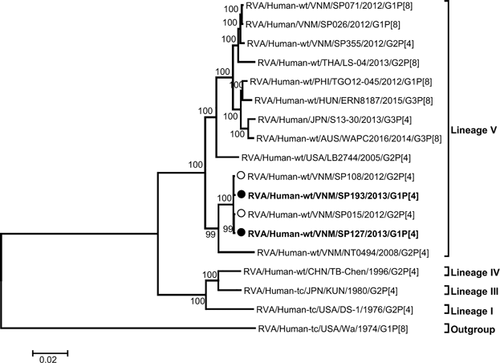
Additional evidence in support of the possible parental source of the genes of the G1P[4] mono-reassortant was provided by phylogenetic analysis. The G1 VP7 genes of SP127 and SP193 were found to be closest to SP110, a Vietnamese G1P[8] strain that had co-circulated during the study period (Fig. 2), whereas their P[4] VP4 genes were closest to those of the G2P[4] strains, SP015 and SP108, that had co-circulated during the study period in Vietnam (Fig. 3). The topology of the phylogenetic tree of the concatenated VP6, VP1–VP3 and NSP1–NSP5 open reading frames (Fig. 4) revealed a close ancestral relationship between our study strains and those of the co-circulating Vietnamese G2P[4] strains SP015 and SP108 in these genome segments. This observation was consistent with the high nucleotide sequence identities found when the genes were compared individually. Taken together, these results provide evidence that the G1P[4] mono-reassortant strains were generated locally in Vietnam from co-circulating strains through an inter-genotype reassortment event.
DISCUSSION
Genetic reassortment in rotaviruses is often named as one of the mechanisms by which rotaviruses evolve to introduce genetic diversity into their genome 25. Rotaviruses with unusual genotypes have been hypothesized to be reassortants; however, evidence showing precisely that the parental strains were co-circulating with such reassortants within the same place and period has seldom been provided.
We analyzed the whole genomes of G1P[4] strains suspected to be inter-genotype reassortant RVA strains on the basis of unusual G/P type combinations and the short RNA electropherotype associated with the G1 genotype. Our analysis, which included the whole genome sequence data obtained in our preceding study 16, provides clear, robust molecular evidence for the hypothesis that the two G1P[4] RVA strains detected in Hanoi, Vietnam in the 2012/2013 rotavirus season were generated by genetic reassortment between G1P[8] and G2P[4] RVA strains, both of which co-circulated in the same city during the same rotavirus season.
To the best of our knowledge, there is no precedent for parental strains being identified among locally, co-circulating strains that likely donated the genes carried by any inter-genogroup reassortant strains. In a recent study 16, we reported high nucleotide sequence identities in the backbone genes of a G2P[4] strain (SP355, a minority circulating strain during that study period) and G1P[8] double-gene DS-1-like-reassortant strains. This observation led us to conclude that a locally co-circulating SP355-like G2P[4] strain had donated the DS-1-like backbone genes to generate G1P[8] double-gene reassortants strains in Hanoi, Vietnam. It was, however, surprising that G1P[8] strains showing this level of high sequence identities in both the VP7 and VP4 genes were not found in the same country, Vietnam.
Similarly, in another study conducted in Japan 17, we identified a minor G2P[4] strain (88H449) that shared high nucleotide sequence identities with the ten genes of a G1P[4] single-gene reassortant strain (AU64). Our interpretation of this observation was that a locally co-circulating 88H449-like G2P[4] strain had donated the DS-1-like backbone genes to generate the single-gene G1P[4] reassortant, AU64. However, the origin of the G1 VP7 gene was unclear.
We initially suspected that the G1P[4] single-gene reassortants had served as intermediates in the generation of the Vietnamese DS-1-like G1P[8] strains that accounted for 14% of RVA-positive specimens 16. However, our analysis showed that the G1P[4] single-gene reassortants were much less related to the G1P[8] double-gene reassortants detected in the same study site and period.
It is noteworthy that in recent years mono- and double-gene reassortant RVA strains possessing the Wa-like outer capsid genes (G1P[8], G1P[4], G3P[8]) and DS-1-like backbone genes emerged in the Asia-Pacific region, Spain and Hungary 13-17, 26-29. However, the implications of the surge in detection of such atypical RVA strains is unclear. Nonetheless, it is important to monitor the spread of such atypical strains and perhaps investigate whether the virus is likely to make any fitness gains as a result of such inter-genogroup reassortment events.
In conclusion, our findings provide clear, robust molecular evidence that the G1P[4] single-gene reassortants were generated by reassortment between locally co-circulating strains possessing two different genotype constellations.
ACKNOWLEDGEMENTS
We acknowledge the considerable support of the Program for Nurturing Global Leaders in Tropical and Emerging Communicable Diseases, Graduate School of Biomedical Sciences, Nagasaki University. This study was supported in part by a grant from the Japan Agency for Medical Research and Development (AMED: 15fm0108001h001)
DISCLOSURE
The authors have no conflicts of interest to declare.