Unconjugated interferon-stimulated gene 15 specifically interacts with the hepatitis C virus NS5A protein via domain I
ABSTRACT
Interferon-stimulated gene 15 (ISG15), a ubiquitin-like protein, is induced by type I INF. Although several groups have reported ISGylation of the HCV NS5A protein, it is still unclear whether ISGylation of NS5A has anti- or pro-viral effects in hepatitis C virus (HCV) infection. In the present study, the role of ISGylation-independent, unconjugated ISG15 in HCV infection was examined. Immunoprecipitation analyses revealed that ISG15 interacts specifically with NS5A domain I. ISG15 mutants lacking the C-terminal glycine residue that is essential for ISGylation still interacted with NS5A protein. Taken together, these results suggest that unconjugated ISG15 affects the functions of HCV NS5A through protein–protein interaction.
List of Abbreviations
-
- Aa
-
- amino acid
-
- CHIKV
-
- Chikungunya virus
-
- HCV
-
- hepatitis C virus
-
- ISG15
-
- interferon-stimulated gene 15
-
- NS5A
-
- nonstructural protein 5A
-
- pAb
-
- polyclonal antibody
-
- PLA
-
- proximity ligation assay
Hepatitis C virus, a major cause of chronic liver disease, affects 130–170 million people worldwide 1. Chronic HCV infection causes hepatic steatosis, fibrosis, cirrhosis and hepatocellular carcinoma 2. HCV belongs to the Hepacivirus genus of the Flaviviridae family. The positive sense single-strand RNA genome encodes a polyprotein that is cleaved by cellular signalases and viral proteases into 10 viral proteins: three structural proteins (core, E1 and E2), a p7 polypeptide and six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) 3. Unlike other HCV non-structural proteins such as viral protease or polymerase, HCV NS5A lacks any enzymatic motif. It has been well established that NS5A is an essential viral component for efficient viral replication through recruitment of and interaction with various cellular host factors 4.
Interferon-stimulated gene 15 is a type I IFN-inducible gene that encodes for a ubiquitin-like protein containing two ubiquitin-like domains 5. Because the modification of protein by ISG15 is similar to that by ubiquitylation, protein conjugation by ISG15 is termed ISGylation. ISGylation requires three distinct enzymes: an adenosine 5'-triphosphate-dependent activating enzyme for ISG15 (E1, UbE1L), an ISG15-specific carrier protein/conjugating enzyme (E2, UbcH8) and several ISG15-specific conjugating enzymes (E3, Herc5). Expression of ISG15 and conjugation of ISG15 to target proteins are strongly promoted by IFN-α/β treatment, dsRNA and viral or bacterial infection 6. In addition to existing in its conjugated form, ISG15 is present intracellularly in an unconjugated form and is also released into the extracellular space 7.
Hepatitis C virus infection triggers intrahepatic synthesis of several ISGs in humans 8, 9. ISG15, one of the ISGs that are strongly induced by HCV infection 10, has previously been shown to function as an antiviral factor against numerous virus infections, but to act as a proviral factor in HCV infection 11-14. Conversely, other groups have reported that ISG15 mediates antiviral activity against HCV infection, and that this activity may occur through the proteasomal degradation pathway and in a Herc5-independent manner 15, 16. Therefore, the potential roles of ISG15 in HCV infection are still under debate. Evidence is accumulating that unconjugated ISG15 may modulate viral infection independent of ISGylation 17-19. In the present study, we attempted to gain further insight into the role of ISGylation-independent, unconjugated ISG15 in HCV infection.
First, we sought to determine whether ISG15 interacts physically with any HCV proteins in Huh-7 cells. For this purpose, Huh-7 cells were co-expressed with a series of myc-His6-tagged HCV proteins together with HA-ISG15 (Fig. 1). Immunoprecipitation analysis revealed that NS5A-myc-His6 co-immunoprecipitated with HA-ISG15 (Fig. 1a, upper panel, Lane 3), whereas FLAG-ISG15 co-immunoprecipitated with HA-NS5A (Fig. 1b, upper panel, Lane 3). Although only NS3-myc-His6 co-immunoprecipitated with HA-ISG15 (Fig. 1a, upper panel, Lane 5), HA-NS3 did not co-immunoprecipitate with FLAG-ISG15 (Fig. 1b, upper panel, Lane 5) when NS3 was expressed as NS3/4A in the cells (Fig. 1b). NS4A is known to be responsible for the subcellular localization of the NS3-4A complex at the organelle's membranes 20. Thus, we concluded that NS3 may not interact with ISG15 in the appropriate subcellular localization. In addition, neither myc-His6-tagged NS4B, NS5B or p7 protein co-immunoprecipitated with HA-ISG15 (Fig.1a, upper panel, Lane 7; Fig. 1c, upper panel, Lane 5; Fig. 1e, upper panel, Lane 3). As shown in Figure 1d, HCV core protein did not co-immunoprecipitate with HA-ISG15 (Fig. 1d, upper panel, Lane 3). These results suggest that ISG15 specifically interacts with HCV NS5A protein.
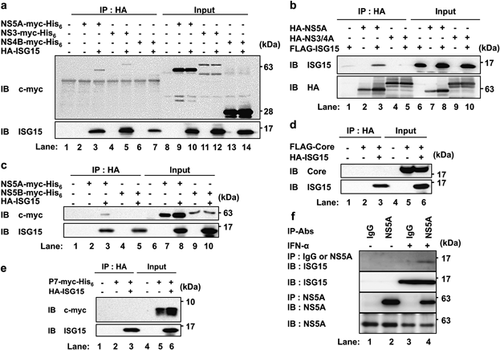
To determine whether endogenous ISG15 interacts with HCV NS5A in HCV-replicating cells, HCV subgenomic replicon cells, Huh 9–13 21 were treated with 1.5 × 103 IU/mL IFN-α (Sigma–Aldrich, St. Louis, MO, USA) for 18 hr. Endogenous ISG15 was co-immunoprecipitated with NS5A protein using anti-NS5A mAb but not with control mouse IgG (Fig. 1f, top panel, Lanes 4 and 3). These results suggest that endogenous ISG15 indeed interacts with HCV NS5A protein expressed by HCV-replicating cells.
To determine whether the interaction between unconjugated ISG15 and HCV NS5A protein is ISGylation-independent, ISG15 mutants with amino acid substitutions in the C-terminal LRLRGG motif (mutated from LRLRGG to LRLRAA, referred to as ISG15 G156/157A) or deleting LRLRGG motif (referred to as ISG15 Δ152-157) were constructed (Fig. 2a). These two ISG15 mutants are incapable of forming ISG15-conjugates 22.
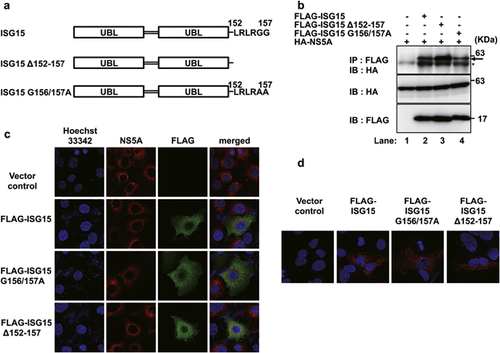
To determine whether these two ISG15 mutants can interact with NS5A protein, HA-NS5A was co-expressed with either FLAG-ISG15, FLAG-ISG15 Δ152-157 or FLAG-ISG15 G156/157A in Huh-7 cells. The cell extracts were subjected to immunoprecipitation with anti-FLAG mAb or anti-HA pAb. As shown in Figure 2b, HA-tagged NS5A co-immunoprecipitated with FLAG-ISG15 Δ152-157 and FLAG-ISG15 G156/157A (Fig. 2b, top panel, Lanes 3 and 4). These results suggest that unconjugated ISG15 interacts with HCV NS5A in an ISGylation-independent manner.
To examine the subcellular localization of ISG15 and NS5A using a confocal microscope, the HCV 1b full-length replicon (RCYM1) 23 was expressed with a vector control, FLAG-ISG15, FLAG-ISG15 Δ152-157 or FLAG-ISG15 G156/157A. Immunofluorescence staining revealed that unconjugated ISG15 colocalized with NS5A in the perinuclear regions of HCV replicon cells (Fig. 2c, merged panels). Moreover, to verify the colocalization of ISG15 and NS5A, an in situ PLA was performed using Duolink In Situ PLA (Sigma–Aldrich) 24. Red fluorescent spots were observed in HCV replicon cells expressing FLAG-ISG15, FLAG-ISG15 G156/157A or FLAG-ISG15 Δ152-157, whereas no signal was detected in the vector control (Fig. 2d). These results suggest that unconjugated ISG15 colocalizes with HCV NS5A protein in an ISGylation-independent manner.
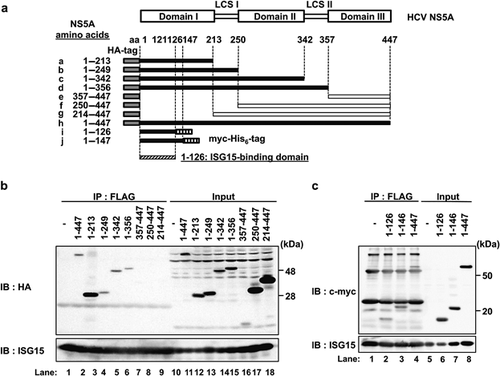
Finally, a previously constructed series of NS5A-deletion mutants were used to determine the ISG15-binding site on NS5A protein 25, 26. The series of NS5A-deletion mutants were co-expressed with FLAG-ISG15 in Huh-7 cells (Fig. 3a). Immunoprecipitation analysis was performed with anti-FLAG antibody, followed by immunoblotting with anti-HA PAb, anti-Myc MAb or anti-ISG15 MAb (Fig. 3b,c). Of the HA-tagged NS5A mutants shown in Figure 3a, N-terminal-deletion mutants lacking the region from aa 357–447, aa 250–447 or aa 214–447 did not co-immunoprecipitate with FLAG-tagged ISG15 (Fig. 3, upper panel, Lanes 7–9), whereas other C-terminal-deletion mutants did co-immunoprecipitate with FLAG-tagged ISG15 (Fig 3b, upper panel, Lanes 2–6). Co-immunoprecipitation analysis using Myc-His6-tagged NS5A 1-126, NS5A 1–146 or NS5A 1–447 revealed that these NS5A mutants co-precipitated with FLAG-ISG15 (Fig. 3c, upper panel, Lanes 2–4). These results suggest that the region from aa 1–126 within the domain-I of NS5A is responsible for the interaction with unconjugated ISG15.
Taken together, these findings suggest that unconjugated ISG15 interacts with NS5A in transfected cells and HCV-replicating cells. Moreover, immunofluorescence staining and in situ PLA assay also demonstrated colocalization of unconjugated ISG15 with the HCV NS5A protein in an ISGylation-independent manner. The region ranging from aa 1 to 126 within the domain I of NS5A, which is the region found to be responsible for the interaction between unconjugated ISG15 and NS5A, was mapped. As far as we know, this is the first report to show a potential role of unconjugated ISG15 in HCV infection.
After discovery of ISG15, much effort was invested in studying ISGylation. Recently, it was reported that ISG15−/− mice are more susceptible to CHIKV infection than are wild-type mice, whereas UbE1L−/− mice display similar susceptibility to CHIKV as wild-type mice, suggesting that ISG15 has a conjugation-independent role in CHIKV infection 17. Other studies have also shown that unconjugated ISG15 interacts with E3 ubiquitin ligase Nedd4 and blocks Nedd4-mediated ubiquitylation of Ebola virus VP40 for efficient release of viral particles 18, 19. These results may imply that viral infection is modulated through unconjugated ISG15. Our results raise the possibility that unconjugated ISG15 has some roles in the HCV life cycle and in viral pathogenesis.
HCV NS5A, a proline-rich, hydrophilic phosphoprotein, functions as a key regulator of HCV RNA replication and assembly 27. NS5A can interact with a wide variety of host cell proteins, including hepatocyte nuclear factor 1α, apolipoprotein A1, 2′-5′oligoadenylate synthase, La, heat shock protein 27, PKR, FK506-binding protein and ovarian tumor protein 7B 25, 26, 28-34. Here, we demonstrated a specific interaction between unconjugated ISG15 and NS5A via domain I of NS5A (Fig. 3). Elucidation of the physiological significance of unconjugated ISG15-NS5A interaction, will require determination of whether unconjugated ISG15 is involved in viral replication, assembly or immune evasion from the host innate immune response. Our results suggest that both unconjugated ISG15 and ISGylation forms of NS5A are present in HCV-replicating cells. Co-existence of unconjugated- and conjugated-ISG15 interactions with NS5A may affect versatile functions of NS5A protein in HCV replication and pathogenesis. Further analysis is required to elucidate the biological significance of the interplay between unconjugated- and conjugated-ISG15.
Collectively, we have here provided evidence that unconjugated ISG15 interacts with HCV NS5A in an ISGylation-independent manner. Understanding the role of unconjugated ISG15 in HCV infection may shed new light on the molecular mechanisms of the HCV life cycle and HCV pathogenesis.
ACKNOWLEDGMENTS
We thank Ms. Yasuko Kozaki for secretarial work. This study was supported in part by grants-in-aid for research on hepatitis from the Ministry of Health, Labour, and Welfare, Japan, and the Ministry of Education, Culture, Sports, Science and Technology, Japan, Japan Society for the Promotion of Science (KAKENHI) and the Japan Agency for Medical Research and Development.
DISCLOSURE
The authors declare that they have no conflicting interests.