Mycoplasma bovis isolates from dairy calves in Japan have less susceptibility than a reference strain to all approved macrolides associated with a point mutation (G748A) combined with multiple species-specific nucleotide alterations in 23S rRNA
ABSTRACT
Erythromycin, tylosin and tilmicosin are approved for use in cattle in Japan, the latter two being used to treat Mycoplasma bovis infection. In this study, 58 M. bovis isolates obtained from Japanese dairy calves all exhibited reduced susceptibility to these macrolides, this widespread reduced susceptibility being attributable to a few dominant lineages. All 58 isolates contained the G748A variant in both the rrl3 and rrl4 alleles of 23S rRNA, whereas a reference strain (PG45) did not. G748 localizes in the central loop of domain II (from C744 to A753) of 23S rRNA, which participates in binding to mycinose, a sugar residue present in both tylosin and tilmicosin. A number of in vitro-selected mutants derived from M. bovis PG45 showed reduced susceptibility to tylosin and tilmicosin and contained a nucleotide insertion within the central loop of domain II of rrl3 (U747–G748Ins_CU/GU or A743–U744Ins_UA), suggesting that mutations around G748 confer this reduced susceptibility phenotype. However, other Mycoplasma species containing G748A were susceptible to tylosin and tilmicosin. Sequence comparison with Escherichia coli revealed that M. bovis PG45 and isolates harbored five nucleotide alterations (U744C, G745A, U746C, A752C and A753G) in the central loop of domain II of 23S rRNA, whereas other Mycoplasma species lacked at least two of these five nucleotide alterations. It was therefore concluded that G748 mutations in combination with species-specific nucleotide alterations in the central loop of domain II of 23S rRNA are likely sufficient to reduce susceptibility of M. bovis to tylosin and tilmicosin.
List of Abbreviations
-
- MIC
-
- minimum inhibitory concentration
-
- RAPD
-
- random amplified polymorphic DNA
-
- rrnA
-
- A2058G mutation in the rrl3 operon
-
- TMS
-
- tilmicosin
Some species of Mycoplasma, the smallest free-living organisms, are pathogenic in food-producing animals 1. Mycoplasma bovis is a causative agent of bovine respiratory diseases, mastitis and arthritis, causes considerable morbidity and has a substantial negative economic effect on the cattle industry 2. Antimicrobials such as fluoroquinolones, macrolides and tetracyclines are key agents for treating M. bovis infections and for preventing their spread in dairy calves. In Japan, three macrolides (erythromycin, tylosin and tilmicosin) are currently approved for use in cattle. Erythromycin contains a 14-membered lactone ring, whereas tylosin and its derivative tilmicosin contain 16-membered lactone rings 3. Tylosin and tilmicosin are commonly used to treat M. bovis infection in cattle, whereas erythromycin is not. On the basis of its 16S rRNA sequence, M. bovis belongs phylogenetically to the Mycoplasma hominis group 4. M. hominis shares a species-specific genetic background for intrinsic resistance to erythromycin; namely, a G2057A substitution in domain V of 23S rRNA 5. M. bovis PG45, the entire genome of which has been sequenced 6, contains this substitution.
The MICs of tylosin and tilmicosin against M. bovis range from 0.5 µg/mL to >512 µg/mL worldwide 7-14. Having less susceptibility to tylosin and tilmicosin than wild-type M. bovis correlates with the presence of one or two point mutations (G748A, C752T, A2058G, A2059G or A2059C, according to the numbering of the Escherichia coli sequence) in the 23S rRNA alleles rrl3 and rrl4 in M. bovis isolates 15. The A2058G mutation in the rrl3 operon (denoted as rrnA in this article) is associated with reduced susceptibility to both macrolides and lincosamides 16. These findings are consistent with those of genetic studies of susceptibility to tylosin and tilmicosin in other Mycoplasma species, and with the results of basic studies in E. coli 17-19. However, further investigation is required to clarify whether these mutations cause less susceptibility than a reference strain to macrolides in Mycoplasma. Because no ‘gold standard’ direct genetic examination has been available, the biological relevance of genetic mutations that are associated with macrolide resistance in Mycoplasma species is typically determined by in vitro selection of mutants that show little susceptibility, with genomic alignment to corresponding E. coli nucleotide sequences 5, 20-22. Such evaluations have not previously been performed for M. bovis. Investigation of molecular mechanisms underlying less susceptibility than a reference strain to macrolides could provide valuable information to guide strategies for treating M. bovis infections and preventing the spread of resistant M. bovis in cattle.
In this study, the contribution of mutations in the genes encoding 23S rRNA and the ribosomal proteins L4 (RplD) and L22 (RplV) to susceptibility to tylosin and tilmicosin was investigated in M. bovis isolates from dairy calves in Japan. M. bovis mutants with less susceptibility to macrolides than wild-type strains were generated by in vitro selection and characterized. Susceptibility to lincomycin, which has not been approved for use in cattle in Japan, was also investigated. Lincomycin has a similar mode of action to macrolides, but differs structurally. Our results suggest that genetic alterations in M. bovis that combine to confer reduced susceptibility to all three approved macrolides, but not to lincomycin, are widely prevalent.
MATERIALS AND METHODS
Bacterial strains
Analyses were performed on 58 M. bovis isolates that had previously been obtained from nasal swabs of dairy calves from 51 farms (each with ≥100 cattle) located in seven regions of Hokkaido Prefecture in Japan (Dounan, Iburi–Hidaka, Ishikari–Sorachi, Kitami–Okhotsk, Kushiro–Nemuro, Soya–Rumoi–Kamikawa and Tokachi) in 2010–2011 23.
These regions cover ∼83,450 km2, and include most of the major dairy-producing areas of Japan. The M. bovis had been isolated and identified previously 23, 24, and then stored until use at −80°C in NK broth (Kanto Chemical, Tokyo, Japan) after cultivation at 37°C in 5% CO2 for 3 days. M. bovis isolates had been spread on modified Hayflick agar (35.5 g/L pleuropneumonia-like-organism agar base, 15% [v/v] horse serum, 2.5% [w/v] yeast extract, 0.0024% [w/v] deoxyribonucleic acid) (Kanto Chemical) from the stock and cultured at 37°C in a 5% CO2 atmosphere for 3 days. Colonies were picked up, inoculated into 1 mL NK broth, and cultured at 37°C in a 5% CO2 atmosphere for 72 hr for antimicrobial susceptibility testing and genetic analysis. Permission had been obtained from all farmers to collect the specimens. The experiments were performed using bacterial strains isolated as part of routine veterinary examination and did not involve any invasive procedures.
Susceptibilities to macrolides and lincomycin
MICs were determined by the agar-dilution method, with modified Hayflick agar and 104–105 CFUs per plate, as described previously 23. The MIC was defined as the lowest concentration of antibiotic that completely inhibited colony formation at 37°C in a 5% CO2 atmosphere following a 72 hr incubation 23, 25. MIC breakpoints for M. bovis have not yet been defined 26, strains with high MICs (≥64 µg/mL) were therefore defined as having les susceptibility than a laboratory strain, M. bovis PG45 (ATCC25523).
RAPD PCR
Genomic DNA of M. bovis isolates was extracted with a Wizard Genomic DNA Purification Kit (Promega, Fitchburg, WI, USA) from the bacteria in 3 mL of Mycoplasma NK broth after 5 days of culture. RAPD PCR was performed using a KAPA2G Fast HotStart MultiPlex PCR kit (Kapa Biosystems, Wilmington, MA, USA) and the primer Hum4 27. The 25 µL PCR mixture contained 10 ng of template DNA, 1 × KAPA2G Buffer A, 0.2 mM of each dNTP, 0.8 µM Hum4 and 0.5 U of KAPA2G Fast HotStart DNA polymerase. The PCR protocol was as follows: 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 36°C for 15 s, and extension at 72°C for 20 s and a final extension at 72°C for 7 min. PCR was performed in a GeneAmp PCR System 2400 (PerkinElmer, Waltham, MA, USA), and 5 µL of amplified products was separated by electrophoresis on 1.5% (w/v) agarose gels in 1 × Tris–acetate–EDTA buffer, followed by staining with ethidium bromide for visualization by ultraviolet transillumination. RAPD profiles were digitized for analysis by BioNumerics version 5.10 software (Applied Maths, Sint-Martens-Latem, Belgium). Similarity coefficients were calculated with the Dice algorithm. Clustering was performed by the unweighted pair-group method with arithmetic mean algorithm. The tolerance and optimization levels were 2.0%.
In vitro selection of M. bovis mutants with reduced susceptibility to tylosin and tilmicosin
M. bovis PG45 was cultured in 10 mL of Mycoplasma NK broth for 3 days and then centrifuged at 10,000 rpm for 10 min. Cell pellets were resuspended in 1 mL of 0.85% (w/v) NaCl and then spread (∼1010 CFU) onto modified Hayflick agar containing 8 µg/mL tylosin or 32 µg/mL tilmicosin (concentrations fourfold higher than the MICs). Cells were cultured at 37°C in a 5% CO2 atmosphere for 1 week, after which colonies that had grown were subcultured at least twice on agar containing tylosin (8 µg/mL) or tilmicosin (32 µg/mL).
Sequencing of 23S rRNA genes (rrl3 and rrl4) and ribosomal protein genes (rplD and rplV)
Nucleotide sequences of rrl3 and rrl4, rplD, and rplV were determined by PCR and direct DNA sequencing. PCR was performed using AmpliTaq Gold (Thermo Fisher Scientific, Waltham, MA, USA) with the primer pairs listed in Table 1. The PCR primers were designed according to the nucleotide sequence of M. bovis PG45 (GenBank accession number NC_014760). The PCR mixture (50 µL) contained 20 ng of template DNA, 1 × buffer I., 5 mM of MgCl2, 0.25 mM of each dNTP, 0.8 µM of each primer and 1.25 U of AmpliTaq Gold DNA polymerase. The PCR protocol was as follows: 94°C for 10 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 2 min, extension at 72°C for 2 min and a final extension at 72°C for 7 min. PCR was performed in a GeneAmp PCR System 2400. The PCR products were purified by using Wizard SV Gel and PCR Clean-Up system according to the manufacturer's instructions (Promega). Nucleotide sequences of the amplified genes were determined using a BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and a 3130 Genetic Analyzer (Thermo Fisher Scientific), with M. bovis PG45 as a reference strain. Accuracy of the prediction of 23S rRNA secondary structure was determined using a bioinformatics tool, CentroidFold 28, 29. Sequences were converted to the E. coli nucleotide numbering by reference to E. coli strain K12 substrain MG1655 (accession number U00096). M. bovis PG45 was the reference for numbering the amino acid substitutions of L4 and L22.
| Primer | Sequence (5′–3′) | Position† | Purpose |
|---|---|---|---|
| MBOVPG45_0282-F | ACTTATCTCTAAATTCAAAAGGATATCTAAC | −2209 bp to +399 bp‡ | DNA amplification of rrl3 |
| 23S-5R | GCCAGGATCAAACTCTCGAAAAA | ||
| 23S-1F | AGAACGTGGGGATGGATTACCTC | −338 bp to +453 bp§ | DNA amplification of rrl4 |
| secY-R | TTAACTGTAGTCATAATAAGGTAGATTGC | ||
| 23S-1R | TCTCTACGGCTCCGCTTTTATCTG | 647–670 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-2F | GAAAGGTGAAAAGAACCCCGAGA | 508–530 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-2R | CATTGCCCTAAGAACGCTCCTCT | 1219–1241 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-3F | TCAGCTAAGGTCCCAAAATCGTG | 1032–1054 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-3R | TCGCATAAGCTAACGCATCCACT | 1852–1874 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-4F | AAATGACCCCGTAAGTTCGCAAG | 1700–1722 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-4R | GAACCGACTCCAGCTCCAGGAT | 2515–2537 bp¶ | Sequencing of rrl3 and rrl4 |
| 23S-5F | GGCAGTCGCTCAACGGATAAAAG | 2412–2434 bp¶ | Sequencing of rrl3 and rrl4 |
| Mb.rplD-F | GCTGAACAAGCTGCACAAGC | −56 bp to +62 bp†† | DNA amplification and sequencing of RplD gene encoding L4 protein |
| Mb.rplD-R | CCTAAGGCATCATTTGACTTTTCA | ||
| Mb.rplV-F | CAACGGTCGTCAACACATCG | −123 bp to +39 bp‡‡ | DNA amplification and sequencing of RplV gene encoding L22 protein |
| Mb.rplV-R | CCATAACGGAAGCCATTTGGA |
- †Nucleotide positions of the 23S rRNA operons (rrl3 and rrl4) are based on M. bovis PG45 numbering (accession no. NC_014760.1); ‡nucleotide positions upstream and downstream of rrl3; §nucleotide positions upstream and downstream of rrl4; ¶nucleotide position within rrl3 or rrl4; ††nucleotide positions upstream and downstream of rplD; ‡ ‡nucleotide positions upstream and downstream of rplV.
The 23S rRNA nucleotide-alteration genotypes of M. bovis were compared with those of other Mycoplasma species for which the MICs of tylosin and tilmicosin have been determined. These strains are M. hominis PG21 (accession number NC_013511.1), M. hyopneumoniae J (accession number NC_007295), M. hyorhinis BTS7 (accession number AB182581), M. pneumoniae FH (accession number NC_017504.1) and M. putrefaciens KS1 (accession number NC_015946.1).
RESULTS
Susceptibilities to macrolides and lincomycin and 23S rRNA genotyping of M. bovis isolates from dairy calves in Japan
There was no variation in MICs to each of the macrolides and to lincomycin across the M. bovis isolates (58 strains) derived from dairy calves in Japan (Table 2). The MICs were high (≥64 µg/mL) for tylosin and tilmicosin in these isolates, but low in the wild-type PG45 strain (2 µg/mL and 8 µg/mL, respectively). MICs were high for erythromycin in both PG45 (128 µg/mL) and the isolates (≥128 µg/mL), and low for lincomycin (2–16 µg/mL).
| MIC†, μg/mL (MIC range) | 23S rRNA mutations‡ | ||||||
|---|---|---|---|---|---|---|---|
| 23S rRNA mutation genotype | Number of strains | ERY | TYL | TMS | LCM | rrl3 | rrl4 |
| Wild-type | PG45§ | 128 | 2 | 8 | 4 | −¶ | − |
| I | 48 | ≥128 (64 to ≥128) | 64 (64 to ≥128) | ≥128 | 4 (2–16) | G748A | G748A |
| II | 5 | ≥128 | 64 | ≥128 | 4 (4–8) | G748A | G748A, C758U |
| III | 2 | ≥128 | ≥128 | ≥128 | 16 | G748A | G748A, C1905A |
| IV | 3 | ≥128 | 64 (64 to ≥128) | ≥128 | 8 (8–16) | G748A | G748A, A2107U |
- †Values are shown as modes; ‡E. coli numbering is used; §M. bovis PG45 was used as the wild-type strain; ¶−, wild-type. ERY, erythromycin; LCM, lincomycin; TMS, tilmicosin; TYL, tylosin. G748 corresponds to G788 in M. bovis numbering, C758 to C798, C1905 to C1904 and A2107 to C2106.
Four genotypes of 23S rRNA mutations (Types I–IV) were identified in the 58 M. bovis isolates (Table 2). All M. bovis isolates contained a common single-nucleotide mutation, G748A, which was present in both 23S rRNA operons, rrl3 and rrl4. Type I isolates (48 strains) did not harbor any 23S rRNA mutations other than G748A. Among Type I isolates, five strains had mutations encoding the amino acid substitution Q93K in L22 (data not shown). Six Type I strains contained multiple amino acid substitutions in L4 (S18T, T43A, A51T, V69A, A70T, E75A, A86T, A101T, V107S, E109_A110InsVKEV) and in L22 (Q93H). These L4 substitutions differ from those identified previously 15. Isolates of Types II–IV contained additional mutations in rrl4: C758U (Type II), C1905A (Type III) and A2107U (Type IV), but no amino acid substitutions in L4 and L22.
On the basis of the results of RAPD PCR with the primer Hum4, PG45 and the isolates were classified into four clusters (A–D in Fig. 1) with a cut-off value of 80% similarity. Approximately 70% (41/58) of the isolates belonged to the major cluster B, which included isolates with 23S rRNA mutation genotypes I, III and IV. Cluster D contained the wild-type PG45, in addition to all five isolates of genotype II.
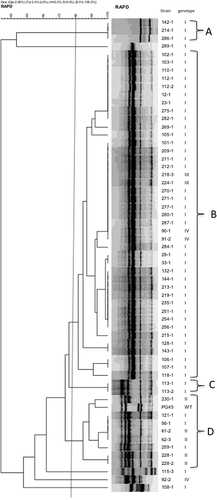
Genotyping of in vitro-selected PG45-derived mutants with reduced susceptibility to macrolides
Twenty mutants were derived by selection by exposure of PG45 to macrolides; 10 each from supplementation with tylosin and tilmicosin (Table 3). The mutants all had nucleotide mutations in rrl3, enabling classification into four genotypes (and a total of seven subgroups) (Table 3). No mutations were present in rrl4. The mutants formed two groups on the basis of susceptibility to macrolides and lincomycin.
| Number of mutants | MIC: μg/mL (MIC range) | 23S rRNA mutations† | |||||
|---|---|---|---|---|---|---|---|
| 23S rRNA mutation genotype | TYL | TMS | TYL | TMS | LCM | rrl3 | rrl4 |
| Wild-type‡ | − | − | 2 | 8 | 4 | −§ | − |
| Va | 0 | 1 | ≥128 | ≥128 | 4 | U747-G748Ins_CU | − |
| Vb | 0 | 4 | 64 | ≥128 | 4 | U747-G748Ins_GU | − |
| VI | 1 | 3 | ≥128 | ≥128 | 4 | A743-U744Ins_UA | − |
| VIIa | 6 | 0 | ≥128 | ≥128 | 4 | A2062U | − |
| VIIb | 2 | 0 | ≥128 | ≥128 | 4 | A2062C | − |
| VIIIa | 0 | 2 | ≥128 | ≥128 | ≥64 | A2058U | − |
| VIIIb | 1 | 0 | ≥128 | ≥128 | ≥64 | A2058C | − |
- †E. coli numbering; ‡M. bovis PG45 was used as the wild-type strain; §−, wild-type. LCM, lincomycin; TMS, tilmicosin; TYL, tylosin; A743 corresponds to A783 in M. bovis numbering, U744 to C784, U747 to U787, G748 to G788, A2058 to A2057, and A2062 to A2061.
One group (Types V–VII) contained nine mutants from tylosin-supplemented agar and eight from tilmicosin-supplemented agar; this group had high MICs for tylosin and tilmicosin (≥64 µg/mL), but not for lincomycin (4 µg/mL). Five genotype V (dinucleotide insertion between U747 and G748) mutants were only obtained from tilmicosin-supplemented agar, four genotype VI (dinucleotide insertion between A743 and U744) mutants were obtained from tylosin and tilmicosin supplementation, and eight genotype VII (single-nucleotide substitution at A2062) mutants were only obtained from tylosin-supplemented agar.
The other group contained three Type VIII mutants with high MICs for tylosin and tilmicosin (≥128 µg/mL) and for lincomycin (≥64 µg/mL). The genotype VIII (single-nucleotide substitution at A2058) mutants were obtained from tylosin and tilmicosin supplementation.
DISCUSSION
All M. bovis isolates (58 strains) examined in this study exhibited less susceptibility than a reference strain to all macrolides (erythromycin, tylosin and tilmicosin) approved for treatment of cattle in Japan; however, they were susceptible to lincomycin, a non-approved lincosamide (Table 2). Similar macrolide-susceptibility patterns have been described in previous studies on large and diverse sets of M. bovis isolates in Japan 7, 10, indicating that the currently approved macrolides are likely to be unsuccessful against M. bovis infection in dairy cattle in Japan. Moreover, we have previously demonstrated that 12% (7/58) of these M. bovis isolates are also resistant to fluoroquinolones 23. These results have implications for future selection of antimicrobials for use in the Japanese dairy industry, which are likely to become progressively more restricted.
The high prevalence of M. bovis isolates exhibiting low susceptibility to tylosin and/or tilmicosin has also been reported in some other countries, including the USA 13, UK 8, Hungary 11 and France 9, whereas ∼50% of isolates are susceptible to tylosin and tilmicosin in Israel, including isolates from domestic cattle and from cattle imported from Europe and Australia 15. Notably, susceptibility of M. bovis to most antibiotics seems to have decreased markedly over the past few decades 9. These findings indicate that M. bovis strains with little susceptibility to macrolides have spread worldwide. We suggest that a small number of dominant lineages of M. bovis with less susceptibility than a reference strain to macrolides have spread throughout the Japanese dairy cattle and now have an extremely high prevalence.
Available evidence suggests that low susceptibility of M. bovis isolates to macrolides with 16-membered-ring structures (including tylosin and tilmicosin) is associated with a G748A mutation in both 23S rRNA operons (rrl3 and rrl4) 15. It also suggests that combinations of other mutations (C752T, A2058G and A2059G/C) with G748A are associated with higher MIC values for both tylosin and tilmicosin than individual mutations 15. In the present study, we detected G748A in both rrl3 and rrl4 in all isolates, suggesting that this mutation has an important role in reducing the susceptibility of M. bovis to tylosin and tilmicosin . By contrast, we do not consider that the observed amino acid substitutions in L4 and L22 or some mutations in rrl4 (encoding C758U, C1905A and A2107U alterations in the RNA) contribute to the low susceptibility to tylosin and tilmicosin because we did not observe these changes in all isolates with high MICs.
Some strains of other Mycoplasma species, such as M. hyopneumoniae J, M. hyorhinis BTS7 and M. hominis PG21, harbor the G748A mutation but remain susceptible to 16-membered-ring macrolides, including tylosin and tilmicosin (Table 4, Fig. 2). In addition, a point mutation at G748 only increases the tylosin MIC 2-fold in E. coli; additional mutations being required for the acquisition of reduced susceptibility to tylosin and tilmicosin 30. These data suggest that the G748A substitution is necessary but not sufficient for reducing susceptibility to tylosin and tilmicosin, and that additional factors contribute to this phenotype in M. bovis isolates.
| Mycoplasma strain | |||||
|---|---|---|---|---|---|
| Nucleotide position† | M. bovis PG45 | M. bovis isolates from Japan | M. hyopneumoniae J M. hyorhinis BTS7 M. hominis PG21 | M. pneumoniae FH | M. putrefaciens KS1 |
| U744 | U744C | U744C | U744C | U744C | U744C |
| G745 | G745A | G745A | – | – | – |
| U746 | U746C | U746C | U746C | – | – |
| G748 | – | G748A | G748A | – | – |
| A752 | A752C | A752C | – | A752C | – |
| A753 | A753G | A753G | A753G | A753G | A753G |
| TYL | S | R | S | S | S |
| TMS | S | R | S | n.d. | n.d. |
- †Nucleotide sequence was compared with that of the E. coli reference strain using the following accession numbers: M. hyopneumoniae J (NC_007295), M. hyorhinis BTS7 (AB182581), M. hominis PG21 (NC_013511.1), M. pneumoniae FH (NC_017504.1) and M. putrefaciens KS1 (NC_015946.1). n.d., not determined; R, reduced susceptibility (MIC ≥64 µg/mL); S, susceptible (MIC ≤32 µg/mL); TMS, tilmicosin; TYL, tylosin. Data for susceptibility were taken from previous studies: M. hyopneumoniea J 39; M. hyorhinis BTS7 21; M. hominis PG21 5; M pneumoniae FH 5 and M. putrefaciens KS1 40.
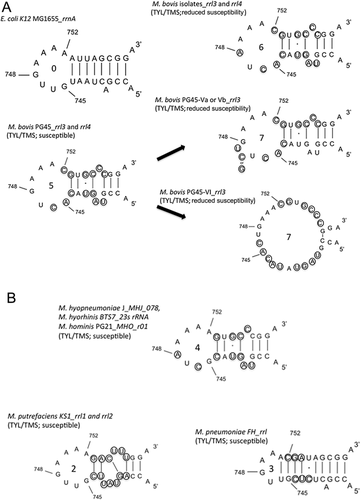
To detect relevant factors other than G748A, we investigated nucleotide alterations in domain II of 23S rRNA of M. bovis (from C744 to G753). This region participates in binding to mycinose, a sugar residue that is present in 16-membered-ring macrolides, such as tylosin and tilmicosin, but absent in other macrolides and lincosamides 3, 31. Nucleotide alterations in this region that suppress interaction with 16-membered-ring macrolides should lead to reductions in susceptibility to them. In the present study, we compared the nucleotide sequences of M. bovis, other Mycoplasma species and E. coli with the aim of identifying any species-specific nucleotide alterations in M. bovis (Table 4). We found that the reference strain PG45 and the M. bovis isolates shared a combination of five nucleotide alterations (U744C, G745A, U746C, A752C and A753G) in the central loop of domain II in both alleles of 23S rRNA that are not present in E. coli (Table 4, Fig. 2). This combination of nucleotide alterations has not previously been reported. We found that other Mycoplasma species lack at least two of the alterations and are susceptible to tylosin and tilmicosin (Table 4, Fig. 2), suggesting that the nucleotide alterations in the central loop of domain II of 23S rRNA are associated with reduced susceptibility to tylosin and tilmicosin. In a previous study, the point mutation G745A occurred during in vitro exposure of M. hyorhinis BTS7 to tylosin, but did not alter the tylosin MIC 21. Although mutation or methylation at G745, G748 or A752 increases tylosin MIC in E. coli, each individual alteration only results in a twofold increase in tylosin MIC 30-32. These lines of evidence suggest that the M. bovis isolates in the present study have evolved reduced susceptibility to tylosin and tilmicosin through occurrence of a common point mutation (G748A) in a background of species-specific nucleotide alterations from the conserved sequence (G745A and A752C, and also perhaps U744C, U746C and A753G) in the central loop of domain II of 23S rRNA. Although we did not fully exclude other factors, such as activation of efflux pumps (which is known to contribute to resistance to other antimicrobials in other Mycoplasma species) 33, our results led us to propose possible molecular mechanisms for the reduced susceptibility of M. bovis to 16-membered-ring macrolides.
Generation and characterization of 20 M. bovis mutants derived from the reference strain PG45 by in vitro selection provided further information on the determinants of antimicrobial susceptibility (Table 3). We found that some of these mutants carry chromosomal alterations that have resulted in nucleotide insertions in the central loop of domain II in rrl3, but not in rrl4, and predicted that these mutations cause drastic changes in the secondary structure of this loop (Fig. 2). These mutations, which contributed to reduced susceptibilities to tylosin and tilmicosin, have not previously been reported in Mycoplasma species. These results provide further evidence that mutations in the central loop of domain II (from A743 to G748) in at least one 23S rRNA allele can lead to reduction in tylosin and tilmicosin susceptibilities.
We found that other mutants generated by in vitro selection harbor mutations at A2062 in rrl3 and exhibit high MICs for tylosin and tilmicosin, but not for lincomycin (Table 3). These mutations have not previously been detected in M. bovis isolates; however, mutations at A2062 contribute to resistance to 16-membered-ring macrolides in M. hominis without affecting susceptibilities to lincomycin and other macrolides 20. These mutations represent an alternative mechanism for reducing susceptibility to 16-membered-ring macrolides; however, that is not currently being observed in M. bovis isolates from dairy cattle in Japan.
We found that a final group of mutants generated by in vitro selection harbor rrl3 mutations at A2058U/C and are associated with high MICs for all the macrolides and lincomycin. A2058, the major binding site for ketolides, macrolides (including those with ring structures containing 14, 15 and 16 members) and lincosamides, forms part of the central loop of domain V of bacterial 23S rRNA 17, 34, 35. Point mutations at A2058 alter the susceptibility of several Mycoplasma species to these agents 22, 36, 37 and A2058G mutations in one or both 23S rRNA alleles have been detected in M. bovis isolates 15, 16. We did not detect A2058G mutations in M. bovis isolates from dairy cattle in Japan in the present study, suggesting that these isolates are still likely to be susceptible to 15-membered-lactone-ring macrolides, ketolides and lincosamides.
Combinations of mutations and nucleotide alterations in 23S rRNA are considered to be associated with reduced susceptibility to macrolides. Point mutations at A2058 in domain V of 23S rRNA of Campylobacter jejuni are associated with low susceptibility to all macrolides, ketolides and lincosamides, but at the expense of a reduction in bacterial fitness 38. G748A mutations in domain II of 23S rRNA may lead to resistance to tylosin and tilmicosin with less effect on fitness than mutations in domain V. M. bovis contains five intrinsic nucleotide alterations (G745A, A752C, U744C, U746C and A753G) in the central loop of domain II of 23S rRNA that are likely to influence binding of tylosin and tilmicosin. Additionally, the G748A mutation could interact with these existing alterations such as to confer resistance to macrolides. The G748A mutation, species-specific nucleotide alterations in the central loop of domain II and the inherited erythromycin-resistant mutation G2057A in domain V of 23S rRNA 5 confer resistance to all macrolides currently approved for use in Japan. The widespread prevalence of low susceptibility to this class of antimicrobial agents has important implications for the treatment of infections in the Japanese dairy industry. Because antimicrobial susceptibility testing is time consuming and difficult for making routine diagnoses in veterinary fields, it may be preferable to institute detection of G748A for rapid estimation of susceptibility of M. bovis isolates to tylosin and tilmicosin. Further study is need to clarify this.
ACKNOWLEDGEMENTS
This study was supported by a grant from a program for developing support systems for upgrading education and research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology and a grant from the Japanese Ministry of Agriculture, Forestry and Fisheries.
DISCLOSURE
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.