Crystal Structure of human pyridoxal kinase: Structural basis of M+ and M2+ activation
Abstract
Pyridoxal kinase catalyzes the transfer of a phosphate group from ATP to the 5′ alcohol of pyridoxine, pyridoxamine, and pyridoxal. In this work, kinetic studies were conducted to examine monovalent cation dependence of human pyridoxal kinase kinetic parameters. The results show that hPLK affinity for ATP and PL is increased manyfold in the presence of K+ when compared to Na+; however, the maximal activity of the Na+ form of the enzyme is more than double the activity in the presence of K+. Other monovalent cations, Li+, Cs+, and Rb+ do not show significant activity. We have determined the crystal structure of hPLK in the unliganded form, and in complex with MgATP to 2.0 and 2.2 Å resolution, respectively. Overall, the two structures show similar open conformation, and likely represent the catalytically idle state. The crystal structure of the MgATP complex also reveals Mg2+ and Na+ acting in tandem to anchor the ATP at the active site. Interestingly, the active site of hPLK acts as a sink to bind several molecules of MPD. The features of monovalent and divalent metal cation binding, active site structure, and vitamin B6 specificity are discussed in terms of the kinetic and structural studies, and are compared with those of the sheep and Escherichia coli enzymes.
Abbreviations: PN, pyridoxine; PL, pyridoxal; PM, pyridoxamine; PLP, pyridoxal 5′-phosphate; ePLK1, E. coli PL kinase1; ePLK2, E. coli PL kinase2; hPLK, human pyridoxal kinase; shPLK, sheep pyridoxal kinase; MPD, 2-methyl-2,4-pentanediol; M+, monovalent cations; M2+, divalent cations.
Pyridoxal 5′-phosphate (PLP) is an essential cofactor serving a vital role in the function of >100 enzymes in amino acid, sugar, and neurotransmitter metabolisms. In many bacteria and plants, PLP is synthesized by a de novo pathway, but most cells rely on a nutritional source of vitamin B6, i.e., pyridoxine (PN), pyridoxal (PL), and pyridoxamine (PM) (Sivaraman et al. 2003). All cells, however, have a salvage pathway for reutilizing PLP liberated during protein turnover (Yang et al. 1996). The salvage pathway involves an ATP-dependent PL kinase that phosphorylates PL, PN, and PM. The product of PN and PM phosphorylation is converted to PLP by pyridoxine 5′-phosphate oxidase, which is also expressed in most cells.
PL kinases have been purified from bacterial, plant, and mammalian sources, and evidence suggests that most eukaryote organisms contain a single PL kinase, coded by a pdxK gene. However, a study with Escherichia coli mutants blocked in the de novo biosynthetic pathway, and with an inactivated pdxK gene that codes for E. coli PL kinase1 (ePLK1), showed that they were still able to grow on PL and led to the discovery of the isoform PL kinase gene, pdxY, which codes for E. coli PL kinase2 (ePLK2) (Yang et al. 1998; Di Salvo et al. 2004). Currently, a large number of prokaryotes expressing PL kinases coded by both pdxK and pdxY genes have been reported. A sequence homology search using the FASTA program (Pearson 1990) found several close homologs of PL kinase, from both prokaryotes and eukaryotes. The sequence identity ranges from 24% to 90% to the human enzyme.
The mechanism of phosphorylation has been elucidated for the sheep and E. coli enzymes, and follows a random sequential substrate addition (Li et al. 2002, 2004; Safo et al. 2004, 2006). In E. coli, the metal ion tandem Mg2+ and K+ are required for enzyme activity (Li et al. 2002; Safo et al. 2006). In contrast, Zn2+ and K+ have been proposed to be the physiological metals needed by both human PL kinase (hPLK) and sheep PL kinase (shPLK) for activity (Li et al. 2002). However, a more recent study with the human enzyme clearly showed that under nonphysiological substrate concentrations and/or at pH 6, where these previous assays were performed, Zn2+ does stimulate the activity (McCormick et al. 1961; White and Dempsey 1970), but under physiological conditions at pH 7.3, Mg2+ is the required divalent metal ion and Zn2+ inhibits the reaction (Di Salvo et al. 2004).
The crystal structures of E. coli PL kinases (ePLK1 and ePLK2) with or without bound substrates have been determined (Safo et al. 2004, 2006). The crystal structures of shPLK alone, and in complex with ZnATP or AMP–PCP/PM or ADP/PLP have also been published (Li et al. 2002, 2004). In addition, the crystal structure of shPLK bound with Roscovitine at the vitamin B6 site is also known (Tang et al. 2005). Recently, a structure of the unliganded human enzyme at a resolution of 2.8 Å has also been reported (Cao et al. 2006).
The effects of several drugs/compounds on PL kinases on mammalian PL kinases have been studied because several important drugs are known to inhibit this enzyme. Some of the earliest studies by the groups of McCormick and Korytnyk showed various analogs of PL and other compounds to be potent inhibitors of PL kinase (McCormick and Snell 1961; Korytnyk and Angelino 1977). Recent studies also show direct inhibitory effects of some drugs on PL kinase, which result in vitamin B6 deficiency and various side effects related to the central nervous system (Hanna et al. 1997; Lainé-Cessac et al. 1997). Well-known examples are theophylline, used to treat asthma, and progabide to treat epilepsy (Lainé-Cessac et al. 1997). Roscovitine, an inhibitor of cyclin-dependent kinases (under study to treat cancers, viral infections, etc.), was recently shown to selectively target and inhibit hPLK (Bach et al. 2005). Some drugs were also found to stimulate the activity of PL kinase and induce depression syndrome (Shetty and Gaitonde 1980).
To characterize the role of the monovalent and divalent cations in the catalytic mechanism of human PL kinase, we performed detailed kinetic and structural studies of hPLK. We also report comparative analyses of the overall protein conformation, active site structures, and kinetic data of the human, sheep, and E. coli enzymes. The structural and functional studies permit investigation at a molecular level to explain why some drugs, such as theophylline, are able to bind to and inhibit this enzyme.
Results and Discussion
Kinetic studies
Our previous studies with ePLK1 showed that K+ was an activator of the enzyme, whereas Na+ did not show any effects nor competed for the K+ binding site (Safo et al. 2006). With hPLK, the effect of monovalent cations is more complicated. We measured enzyme activity using a variety of monovalent cations under saturating PL and MgATP concentrations, and found that both Na+ and K+ activated the kinase when only triethanolamine was present as the cation. Na+ gave a sixfold increase in activity while K+ gave only a 2.5-fold increase (Fig. 1). Double reciprocal plots with varying concentrations of PL and MgATP were linear and crossed on the negative X-axis, suggesting no synergistic binding effect between these two substrates. Under saturating substrate concentrations and 40 mM Na+, kcat was 200 min−1. When the enzyme was present in the K+ form, the kcat was about 85 min−1. The activities of the larger cations, Rb+, Cs+, as well as the smallest cation, Li+ were significantly low compared to Na+ or K+ (data not shown). It is difficult to rule out any contamination of some sodium or potassium in the reagents in the other cations themselves that could account for this low level of activity.
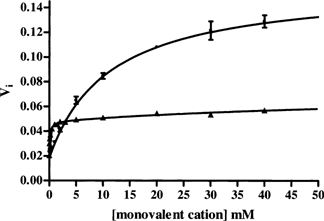
Activity of hPLK in the presence of either Na+ or K+. Monovalent cations were added in increasing concentration to hPLK in buffered triethanolamine BES, pH 7.3 (closed circles for Na+ and triangles for K+). Initial velocity was determined at 388 nm, where the product PLP has it maximal absorbance.
The Km values for PL and MgATP are very different when Na+ or K+ is used as the cation. In the presence of Na+, the Km values were 75 μM and 500 μM for PL and MgATP, respectively. In the presence of K+, the Km values were much lower. We could not obtain accurate initial rates because the substrate concentrations were so low that initial velocity could not be measured accurately. We estimate the Km for PL and MgATP to be lower than 10 μM and 25 μM, respectively (data not shown). Therefore, the affinities of the K+ form of the enzyme for PL and ATP are at least an order of magnitude lower than with the Na+ form of the enzyme.
Both Na+ and K+ showed saturation kinetics (Fig. 1). When Na+ was used as counter cation, there was some inhibition when the concentration was raised above 50 mM. Using initial velocity measurements up to 30 mM results in Na+ exhibiting a Kd value of 12 mM. Although we do not have direct evidence for the cause of this inhibition, it could be the result of ionic strength effects or other Na+ binding sites. Inhibition of enzyme activity by M+ at high concentration has been documented and sometimes attributed to the chaotropic effect of ions that leads to destabilization of the enzyme with a concomitant reduction in catalytic activity (Zhao 2005).
The results for K+ were best fit to two Kd values. The first one gives a Kd of 0.12 mM, and the second one is 80 mM. Again, the higher value may be only an ionic strength effect. Although competitive assays between Na+ and K+ were difficult to perform because of the very different Km values and the similar kcat values, it was clear that these two cations compete for the same site on the enzyme. In the presence of 20 mM Na+ and low concentrations of PL and MgATP, the addition of K+ resulted in increased activity (under these conditions PL and MgATP are saturating for the K+ form of the enzyme but not for the Na+ form). If K+ is added to the more active Na+ form of the enzyme, in the presence of high concentrations of PL and MgATP, inhibition is observed because of the lower kcat value of the K+ form of the enzyme.
Taken together, the above results suggest that under physiological conditions in cells, where K+ is severalfold higher than Na+, the enzyme is in its K+ form and that hPLK should be considered as a K+ activated enzyme. To determine if Na+ is involved under some special circumstances requires further study.
Structure determination and comparison
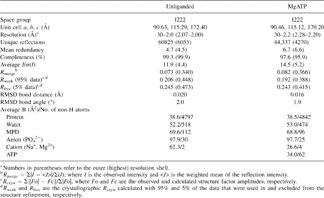
The crystal structures of hPLK and its complex with MgATP are reported here at resolutions of 2.0 Å and 2.2 Å, respectively. The two crystals are isomorphous, and both belong to the orthorhombic space group I222. The unliganded structure was determined using the Web-tool molecular replacement method CaspR (Sali and Blundell 1993; Navaza 1994; Brunger et al. 1998; Notredame et al. 2000; Claude et al. 2004) and refined to a final crystallographic Rwork/Rfree of 20.6% / 24.5%. The structure was subsequently used to refine the hPLK–MgATP complex to crystallographic Rwork/Rfree of 19.2% / 24.3%. The asymmetric unit of both structures contains monomers A and B, which are related by a noncrystallographic dyad axis. Each monomer of hPLK consists of nine α-helices and 11 β-strands (Fig. 2A). The unliganded shPKL, ePLK1, and ePLK2 crystal structures have been reported at 2.1 ∼ 2.2 Å resolution, while the corresponding MgATP complexes of shPKL and ePLK1 have also been reported at 2.6 Å resolution. All these PL kinase structures show the same typical ribokinase superfamily central core of β-sheets surrounded by α-helices, as well as similar dimeric assembly (Mathews et al. 1998; Sigrell et al. 1998; Campobasso et al. 2000; Schumacher et al. 2000; Cheng et al. 2002; Li et al. 2002; Safo et al. 2004, 2006; Zhang et al. 2004; Newman et al. 2006). A structure of the dimer is shown in Figure 2B. The surface area buried at the dimer interface (calculated with the CNS program suite) is 4114 Å2 of the 24,488 Å2 surface areas on both monomers. The corresponding surface area buried values for ePLK1, ePLK2, and shPLK are 3119 Å2, 3410 Å2, and 3860 Å2, respectively, all consistent with dimeric structures. Consistently, mammalian PLK, including that of the human enzyme, functions as a dimer; however, it had been shown to retain catalytic activity upon dissociation into monomers (Kwok et al. 1987).
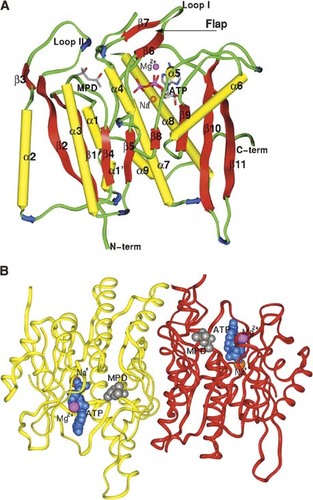
Overall structure of hPLK. (A) The monomeric structure with bound ATP (stick), MPD (stick), Na+ (brown sphere), and Mg2+ (magenta sphere) at the active site. α-Helices and β-strands are colored yellow and red, respectively. The secondary structures are labeled. (B) The dimeric structure, also with bound ATP (cyan CPK), MPD (gray CPK), Na+ (brown sphere), and Mg2+ (magenta sphere). Monomers A and B are colored red and yellow, respectively.
The overall structures of hPLK and hPLK-MgATP are very similar. The dimers (for 577 Cα pair atoms) or monomers (for 299 Cα pair atoms) superimpose on each other with an RMSD of 0.37 Å, which compares with ∼0.35 Å between monomers within each structure. This indicates that binding of ATP at the active site does not induce larger conformational changes than exists between two monomers within the same crystal. Similar observations were also reported for the ePLK1 and shPLK structures (Safo et al. 2006). However, we observe localized but significant differences between the structures at the active site; notably the region of β6- and β7-strand (residues 118–124, including loop I) and in the loop region between strand β11 and helix α7 (residues 224–228) (see Fig. 2A). In fact, both structures have moved significantly closer to the active site from their positions in the unliganded hPLK, apparently due to nucleotide binding, culminating in a more ordered active site structure in the hPLK–MgATP complex.
Not surprisingly, due to the large sequence homology between the eukaryote kinases, comparison between the unliganded hPLK and unliganded shPLK shows a low RMSD of 0.45 Å. A significantly higher RMSD of 0.80 Å was observed between hPLK and the previously published 2.8 Å human kinase structure (Cao et al. 2006). The structural differences are mostly located at the ATP binding site, specifically at the loop/turn between strands β6 and β7; between strand β11 and helix α7; and between strands β9 and β10 (see Fig. 2A). However, the most difference occurs at the turn between strands β10 and β11 (∼3.5 Å shift). The observed structural differences between the two human structures are partly due to the bound MPD in the structure reported here, which has necessitated the displacement of some of these loops or turns. We also note that the 2.8 Å human structure was determined using a de-twinned data set with a completeness of 81% (Cao et al. 2006), and the potential coordinate errors in this structure may also have contributed to the observed structural differences.
In the unliganded hPLK crystal, we observed bound Na+ in both active sites, close to where the ATP γ-phosphate group is located in the hPLK–MgATP complex (see below). The crystallization drop contained a final concentration of 10 mM K+ and 50 mM Na+. The Na+ in monomer A is pentacovalently coordinated with five water molecules (average distance of 2.1 Å). The water molecules are hydrogen bonded to the side chains of Asp113, Glu153, and Thr186, as well as the amide nitrogen and oxygen atoms of Asn150 and Thr148, respectively (Fig. 3A). In monomer B, a similar Na+ configuration is seen, except the fifth water is excluded due to lack of clear electron density. The sodium ions themselves had high temperature factors (Table 1).
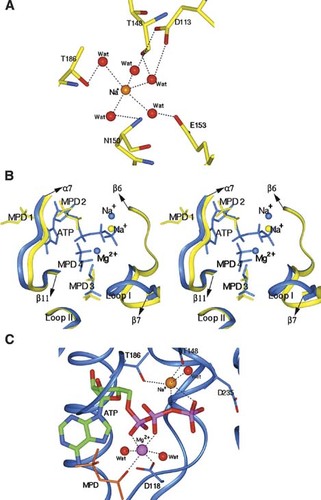
Structural basis of monovalent and divalent ions' activation of hPLK. (A) Na+ (brown sphere) bound to the active site of monomer A of unliganded hPLK. The coordination sphere of Na+ involves only water molecules (red spheres); with the latter mediating intricate hydrogen-bond interactions with the active site protein residues (sticks). (B) Stereoview of the superimposed active sites of unliganded hPLK (colored yellow) and hPLK-MgATP complex (colored cyan). Three MPD molecules occupy the active site of the unliganded hPLK structure, whereas two are found in the hPLK–MgATP complex. Relevant protein side chains, MPD, and ATP are shown in sticks, and the metal ions are shown in spheres. (C) Na+ (brown sphere) and Mg2+ (magenta sphere) binding modes at the active site of hPLK–MgATP complex. The protein backbone is shown in blue ribbons, and relevant protein residues are shown in blue sticks. Both metal ions make contact with the protein, ATP (sticks), water molecules (red spheres), and/or MPD molecule (brown stick).
The hPLK model also contains 14 MPD molecules, located in the active sites, on the surface of the protein at various crevices, as well as the interfaces of crystal contacts. It is noteworthy that binding of MPD has resulted in the crystal diffracting to a high resolution of 2.0 Å despite the high solvent content of 61.4%. Another crystal form of hPLK obtained with ammonium sulfate and without MPD diffracts only to 3.0 Å resolution (data not shown), similar to the recently published 2.8 Å human structure (Cao et al. 2006). The solvent contents of these two latter crystals are 58% and 59%, respectively. Interestingly, three MPD molecules span across the PL (MPD 3) and ATP (MPD 1 and MPD 2) binding cavities in each of the monomers of hPLK, mimicking the substrate molecules (Fig. 3B).
The hPLK–MgATP model contains two ATP molecules, two each of Na+ and Mg2+, five HPO42−, and 12 MPD molecules. Binding of ATP has displaced the bound Na+ in the unliganded hPLK by ∼2.0 Å to another position (Fig. 3B). The Na+ with its coordinating water molecules (as found in the unliganded hPLK) stripped off now makes direct bond contacts with the protein and the ATP (Fig. 3C). Additionally, binding of ATP has also displaced two MPD molecules (MPD 1 and MPD 2 in Fig. 3B) observed at the ATP binding site in the unliganded structure. However, like the unliganded structure, we still observed a MPD molecule bound at the PL binding site (MPD 3 in Fig. 3B). A new MPD molecule (MPD 4 in Fig. 3B) has appeared in the hPLK–MgATP complex, making contact with the bound Mg2+ (see below). These crystallographic observations suggest that the active site of PL kinase may act as a sink to bind to compounds other than the natural substrates. Consistently, functional and/or crystallographic studies indicate that certain drugs and compounds, such as theophylline, progabide, and Roscovitine bind to the active site of PL kinase to inhibit its activity (Hanna et al. 1997; Lainé-Cessac et al. 1997; Bach et al. 2005; Tang et al. 2005). Detailed atomic level studies of these drugs complexed with hPLK using X-ray crystallography are warranted to understand their inhibitory actions. Unlike the clear presence of ATP in the hPLK–MgATP complex, cocrystallization of PL with the protein using MPD did not show PL; neither did the cocrystals of hPLK with AMP–PCP/PL show the bound substrates.
Active site structure
Each of the two active sites in hPLK–MgATP complex binds ATP, Mg2+, and Na+ molecules. Comparison of hPLK and hPLK–MgATP structures shows that residues 224–228, which form part of the loop region between strand β11 and helix α7 have rotated away from the active site to allow binding of the ATP adenine (Fig. 3B). As noted above, binding of the MgATP has also removed two MPD molecules from the ATP binding site (Fig. 3B). Additionally, the active site structure formed by the strand β6-Loop I–strand β7, which is referred to as a flap in PL kinases has moved closer to the bound ATP (Fig. 3B). The structural equivalent of the flap in other members of the ribokinase superfamily of enzymes is referred to as a lid (Mathews et al. 1998; Sigrell et al. 1998; Campobasso et al. 2000; Schumacher et al. 2000; Cheng et al. 2002; Li et al. 2002; Safo et al. 2004, 2006; Zhang et al. 2004; Newman et al. 2006). A key to the random sequential kinetics exhibited by PL kinases is the ability to prevent unproductive hydrolysis of ATP in the absence of a bound vitamin B6, and the flap is believed to play this important role by providing hydrogen-bond interactions to the ATP β- and γ-phosphates (Li et al. 2002, 2004; Safo et al. 2006). Another significant function of the flap is to sequester the ATP for catalysis. So far, the only non-PL kinase of the ribokinase superfamily which appears to have a flap like the PL kinases is the HMPP kinase (Cheng et al. 2002). Detailed analyses of the function and evolutionary trend of the flap/lid in the ribokinase superfamily enzymes have appeared in several publications (Mathews et al. 1998; Sigrell et al. 1998; Campobasso et al. 2000; Schumacher et al. 2000; Cheng et al. 2002; Li et al. 2002; Safo et al. 2004, 2006; Zhang et al. 2004; Newman et al. 2006).
The mode of ATP–protein interaction is depicted in Figure 4A, which is similar to those described for shPLK and ePLK1 (Li et al. 2002, 2004; Safo et al. 2006). However, there are some prominent differences due to variations in the β-/γ-phosphate group positions in the three structures. In particular, the γ-phosphates of shPLK, hPLK, and ePLK1 are about 6.1 Å, 6.8 Å, and 7.6 Å, respectively from the 5′-hydroxyl group of vitamin B6. Thus, the observed protein (Asp118, Asn150, Thr186, Ser187, Asp113, Glu153, Tyr127)-ATP β-/γ-phosphate hydrogen-bond contacts in hPLK are mostly absent in the sheep structure, with the exception of those involving Asp118, Tyr127, and Asn150 (Fig. 4B). Similar differences are also observed with ePLK1. Of note is that all the residues that make contact with the β-/γ-phosphate groups are totally conserved in the three enzymes, and also highly conserved in the rest of the PL kinases. Overall, we see more hydrogen-bond interactions between the ATP and the protein in human than in either the sheep or E. coli structures. We also note that the residues that make van der Waals contacts with the adenine and sugar moieties of ATP are more hydrophobic in the human enzyme. It is therefore not surprising that we observe well-defined ATP electron density in the human structure (Fig. 4C,D), while disorder of the ATP was reported for both the sheep and E. coli structures, especially in the latter structure. We should point out that the ATP-complex structures of ePLK1 and shPLK were reported at a significantly low resolution of 2.6 Å, compared to 2.2 Å for the hPLK–MgATP complex. Note that in the E. coli and sheep structures the B-values for the MgATP or ZnATP atoms (69 and 47 Å2, respectively) were significantly higher than those of the protein only atoms (47 and 32 Å2, respectively) compared to those of the human (protein only = 39 Å2; MgATP only = 34 Å2).
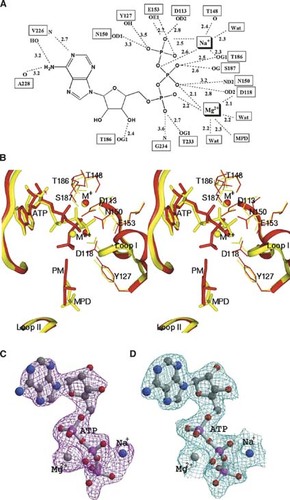
The active site and substrate binding mode of hPLK–MgATP complex. (A) Schematic diagram showing interactions (dotted lines) between Na+, Mg2+, ATP, and the protein residues of hPLK. Bond distances are average of the two active sites. (B) Stereoview of the superimposed active sites of hPLK–MgATP (colored yellow) and shPLK–MgATP (colored red). ATP, PM, and MPD molecules, and relevant protein side chains are shown in sticks. Monovalent ions (Na+ in hPLK and K+ in shPLK) and divalent ions (Mg2+ in hPLK and Zn2+ in shPLK) are shown in spheres. Note that in the hPLK–MgATP structure, MPD is observed bound at the vitamin B6 site, which overlaps the bound PM in the shPLK–MgATP structure. (C) A Fo−Fc map (contoured at the 3.0 σ level) of Na+, Mg2+, and ATP of the hPLK–MgATP complex, calculated before the metal ions and ATP were added to the refined model. (D) A 2Fo−Fc map (contoured at the 1.0 σ level) of Na+, Mg2+, and ATP of the hPLK–MgATP complex. Both maps are superimposed with the final refined models.
Both the unliganded hPLK and the hPLK–MgATP structures show a MPD bound at the PL binding position (noted as MPD 3 in Fig. 3B). Interestingly, when the enzyme was cocrystallized with PL, also using MPD as a precipitant, the difference map shows a density consistent with bound MPD and not PL. The hydroxyl oxygens of the MPD make hydrogen-bond interactions with the main-chain nitrogen and side-chain hydroxyl of Thr47 and Ser12, respectively. Based on the position of the MPD, and the binding mode of vitamin B6 to shPLK and ePLK1, we can readily deduce how PL binds to the active site of the ATP-bound hPLK structure (Fig. 4B). As expected, we observe similar hydrogen-bond and hydrophobic interactions between a modeled PL and the protein as found in shPLK. The residues involved in these interactions (Ser12, Thr47, Asp235, Val19, and Tyr84) are totally conserved, with the exception of Thr47, which is Pro in the E. coli structure.
Central to the function and classification of the PL kinases is another active site structure, Loop II (located between strands β2 and β3; residues 44–54) and its fingerprint a Thr–Gly dipeptide (residues Thr47 and Gly48) (Figs. 2A, 3B, 4B) (Safo et al. 2004, 2006). As previously suggested, Loop II, and in particular the dipeptide motif, has a substrate binding role of specifically sequestering the substrate for catalysis as well as determining substrate affinity and specificity (Li et al. 2002, 2004; Safo et al. 2004, 2006). In eukaryotes, the motif is comprised of Thr47 and Gly48, while in prokaryotes, coded by the PdxY gene, the motif is uniquely Thr–Gln. There are several variations of the motif in prokaryotes coded by the PdxK gene (all distinctly different from the human), with Pro–His occurring most often. In the human and sheep structures, Thr47 makes a hydrogen-bond interaction with the O3′ substituent of PL. Interestingly, as the side chain of residue 48 gets longer, as observed in the E. coli kinases (His in ePLK1 or Gln in ePLK2), it covers the active site from the bulk solvent, and in particular the C4′ position of the substrate. Replacement of His or Gln with Gly, as observed in humans and sheep, leaves the C4′ position uncovered to the bulk solvent and also precludes interaction between the protein and substituents at the C4′ position of vitamin B6. A study by McCormick and Snell, has shown that PL kinases from the rat, humans, and yeast have high tolerance to a wide variation of substitutions at the C4′ position of PL, in contrast to bacterial kinases from Lactobacillus casei and Streptococcus faecalis (McCormick and Snell 1961).
Metal binding and enzyme activity
Metals, both monovalent and divalent cations, are known to be absolute requirements for the mechanistic function of many kinases, providing driving forces for ATP binding and substrate catalysis (Di Cera 2006). A previous study with unpurified human PL kinase from erythrocytes suggested K+ to be the monovalent cation required by this enzyme for activity (Lainé-Cessac and Allain 1996). As noted above, Na+ elicits maximum enzyme activity about twofold higher than K+. On the other hand, both ATP and PL bind with higher affinity in the presence of K+ compared to Na+. Our previous studies with the E. coli kinase found K+ to be the required monovalent cation for enzyme activity (Safo et al. 2006).
In order to determine the positions of the divalent and monovalent metals and to try to understand how these metals are involved in the phosphoryl transfer reaction on a molecular level, we cocrystallized the human kinase with both Mg2+ and Na+. We had previously studied the metal binding sites in the E. coli enzyme by X-ray crystallography (Safo et al. 2006). Nonetheless, the low-resolution ePLK1 crystal structure did not show a clearly defined K+ at the active site, and thus no monovalent ion was included in the structure (Safo et al. 2006). On the other hand, the ATP γ-phosphate group and the two metal ions in the human structure are remarkably well defined in both active sites (Fig. 4C,D). The Mg2+ in hPLK is located between the ATP α- and β-phosphate groups (Figs. 3B,C, 4B). A similar observation is observed in the structure of Toxoplasma gondii adenosine kinase (Schumacher et al. 2000). This is in contrast to the sheep enzyme, where the loosely bound divalent Zn2+ cation is found between the β- and γ-phosphate groups (Fig. 4B). Another significant observation is that Na+ and Mg2+ are situated on the opposite sides of the ATP β-/γ-phosphate groups in hPLK, rather than cis, as found in shPLK (Figs. 3C, 4B). The metal ion tandem in the human structure is Na+ and Mg2+ compared to K+ and Zn2+ in the sheep structure. In hPLK, the Mg2+ is tightly bound with well-defined octahedral coordination (Fig. 3C); two ligands from the ATP α/β-phosphate groups (∼2.1 Å), a ligand each from the carboxyl oxygen of Asp118 (∼2.2 Å) and the hydroxyl of MPD (∼2.3 Å), and two water molecules (∼2.2 Å). The high charge density of Mg2+ is thus balanced by the negatively charged carboxyl and phosphoryl groups, and most importantly the Mg2+ location is consistent with its function by binding to the bis-phosphate moiety to neutralize the negative charges and stabilize the ADP leaving group during the phosphorylation reaction.
The coordination of the Na+ in hPLK-MgATP appears to be trigonal–bipyramidal or square–pyramidal (Fig. 3C), with two donor atoms from the ATP γ-phosphates group (∼2.5 Å), one donor atom from the amide oxygen of Thr148 (∼2.4 Å), and one donor atom from the hydroxyl of Thr186 (∼2.3 Å), and a water molecule (2.3 Å). There are no other protein ligands within 3.4 Å of the Na+ coordination sphere. K+ instead of Na+ was used to crystallize the sheep enzyme; however, the two monovalent metal positions overlap (Fig. 4B). The observation that Na+ and K+ bind at the same site is consistent with our kinetic studies that show competition between Na+ and K+. Nevertheless, while Na+ has a five-coordination shell in the human enzyme, K+ in the sheep enzyme is octahedrally coordinated with the protein residues Thr186, Asp113, Glu153, and Thr148, as well as the ATP β-phosphate group and a water molecule. Thus, when K+ is replaced by Na+ in the human enzyme only the interactions with Thr148 and Thr186 are conserved, and contributions from Asp113 and Glu153 are lost. In the human enzyme, Asp113 and Glu153 are located >3.7 Å from the Na+. Remarkably, the bound Na+ (K+ in sheep) and Mg2+ (Zn2+ in sheep) are separated by 6.8 and 3.4 Å in the human and sheep structures, respectively. However, in both structures, the two metal ions, through their direct interaction with the protein and ATP phosphate groups, should be able to engage and anchor the ATP for catalysis.
The question remains as to why the human enzyme is more active in the presence of Na+ than K+; however, the latter significantly increases the affinity for ATP and seems to be the physiological cation. It is possible that replacement of K+ with Na+ changes the geometry of the active site to a more optimal orientation of catalytic residues, leading to greater enzyme activity in the presence of the later cation. This is consistent with several studies that suggest that coordination sphere of metal cations plays a significant role in enzyme activity. In dialkylglycine decarboxylase, replacement of Na+ with K+ is known to drastically change the geometry of coordination and perturbs residues that control binding of substrate (Toney et al. 1993; 1995). In thrombin, changes in the coordination sphere of Na+ and K+ is known to affect the oxyanion hole geometry explaining the differences in kinetic activity (Pineda et al. 2004; Papaconstantinou et al. 2005). In some proteins, however, replacement of one metal with another, such as K+ with Na+ in pyruvate kinase, results in no structural change even though the enzyme is inactive in the absence of K+ (Larsen et al. 1998).
It should be recalled that in unliganded hPLK, there is a bound Na+, which is penta- or tetra-coordinated with solvent molecules (Fig. 3A). The solvent molecules, in turn, make close and intricate hydrogen bond interactions with the protein residues that help stabilize the active site conformation. Of particular interest is the fact that binding of ATP displaces the Na+ to another position, closer to the protein, where it makes direct interaction with the protein in addition to the ATP (Fig. 3B,C). Note that the B-values of the Na+ ions in the unliganded crystal (∼60 Å2) are much higher than those in the MgATP complex (∼30 Å2), suggesting stable binding with the mediating waters excluded. It is known that substrate binding to a stabilized enzyme–M+ complex is more favorable as the entropic penalty of ordering the enzyme to form the enzyme–substrate complex is offset by the previously bound M+ (Page and Di Cera 2006). This is consistent with the observed increased affinity for ATP in the kinases. Nevertheless, due to the small size of Na+ (0.9 Å), it should form a relatively strong interaction with four or five solvent molecules, while the larger K+ (1.33 Å) should prefer more solvent molecules with relatively weaker interactions (Collins 1995, 1997, 2006; Dill et al. 2005). The ability to strip off water molecules from the hydration sphere should therefore be easier with K+ than with Na+, and could explain why the enzyme shows greater affinity for ATP in the presence of K+ than Na+.
A known K+ position in the E. coli enzyme would help throw light on the structural basis of the M+ selectivity in ePLK1. However, it seems that human and perhaps other eukaryote PL kinases have evolved K+ selectivity by imposing a six-coordinate geometric constraint on the coordination sphere that may not be accessible by Na+. In some ribokinase superfamily enzymes, the monovalent cation does not make contact with the protein but only with the ATP, and it is believed that the cation exerts its influence indirectly by perturbing the conformation of the active site residues (Andersson and Mowbray 2002; Zhang et al. 2004; Di Cera 2006). On the other hand, in some PLP-dependent enzymes, such as serine hydratase (Yamada et al. 2003), tryptophanase (Isupov et al. 1998), and tyrosinase (Sundararaju et al. 1997), the M+ does not make contact with the substrate, but aids to organize the architecture of the active site for catalysis (Di Cera 2006). A recent mini-review article elegantly classified the enzymes that have a monovalent cation bound to ATP and the protein residues as M+-activated Type I, while those that bind only to either the protein or the substrate as M+-activated Type II (Page and Di Cera 2006). PL kinases obviously belong to the former, while other non-PL kinase members in the ribokinase superfamily of proteins, such as aminoimidazole riboside kinase (Zhang et al. 2004) belong to the latter. Thus, the binding mode of monovalent cations may be an indicator as to how these species have evolved from each other.
Materials and Methods
Expression and purification of hPLK
Competent E. coli Rosetta (DE3)pLysS cells (Novagen) were transformed with a pET22b vector carrying the human pdxK gene insert. Transformants were grown in 6 L of LB broth and after reaching an O.D.600nm of 1.2 were induced with 0.5 mM IPTG. Cells were grown for an additional 5 h at 30°C and harvested by centrifugation. The cell pellet was suspended in 200 mL of potassium phosphate buffer, pH 7.2, and ruptured by osmotic shock. Streptomycin sulfate was added to a final concentration of 10 g/L to remove excess nucleic acid. After an ammonium sulfate fractionation, and dialyzing the enzyme against 20 mM potassium phosphate buffer, pH 7.2, it was purified on TMAE, phenyl sepharose, and hydroxyapatite columns, as previously described (Di Salvo et al. 2004). The purity was >95%, as judged by SDS-PAGE.
Kinetic studies
Unless noted otherwise, hPLK used in the kinetic experiments was extensively dialyzed against 20 mM triethanolamine BES [N,N-bis(2-hydroxyethyl)-2-amino-ethanesulfonic acid], pH 7.3, prior to performing assays. All assays were performed at 37°C in a 1-cm thermostated cuvette. Initial velocity for the conversion of PL to PLP was followed at 388 nm in an Agilant 8454 spectrophotometer in 20 mM triethanolamine BES, pH 7.3, and containing added salts as described in the text. The values for Km and kcat were determined from double reciprocal plots. PL concentrations were varied between 10 μM and 300 μM and MgATP concentrations between 100 μM and 1 mM. All reagents used in the assays were dissolved in 20 mM triethanolamine BES, pH 7.3. Dissociation constants for the binding of monovalent cations were calculated from saturation curves of enzyme activity versus cation concentration. Curve-fitting procedures and statistical analysis were carried out using Prism (GraphPad Software).
Crystallization
Human PL kinase protein was dialyzed overnight against 20 mM K phosphate buffer (pH 7.0) containing 100 mM NaCl and then concentrated to 18 mg/mL (0.5 mM). Initial crystallization screening at room temperature by the hanging-drop vapor diffusion technique with either ammonium sulfate or 2-methyl-2,4-pentanediol (MPD) precipitants produced crystals, with the best crystals from MPD. Subsequent crystallization drops composed of 2 μL of protein solution and 2 μL of reservoir and equilibrated against 700 μL reservoir solution (100 mM Tris-HCl, pH 8.0, and 50% MPD) resulted in rectangular-shaped crystals, and reached a maximum size of ∼0.1 × 0.2 × 0.2 mm in about a week. Crystallization of hPLK in the presence of 2 mM MgATP or 1 mM PL or 2 mM AMP–PCP/PL was also attempted under similar conditions. The complex mixtures were first incubated on ice for about 3–4 h before screening for crystallization.
For X-ray data collection, crystals were cryoprotected in solution containing 100 mM Tris-HCl buffer (pH 8.0) and 60% MPD with or without the appropriate substrate (PL or MgATP or AMP-PCP/PL) before flash cooling in a cryogenic nitrogen stream. X-ray data were collected at 100 K using a Molecular Structure Corporation (MSC) X-Stream Cryogenic Crystal Cooler System and an R-Axis IV++ image plate detector, a Rigaku MicroMax–007 X-ray source equipped with MSC Varimax confocal optics operating at 40 kV and 20 mA. Crystals of hPLK in unliganded form diffracted to 2.0 Å; hPLK grown in the presence of MgATP, PL, and AMP–PCP/PL diffracted to 2.2 Å, 2.4 Å, and 2.3 Å, respectively. The data sets were processed with the MSC d*TREK software program. The Matthews Coefficient of 3.2 and water content of 61% are consistent with one dimer per asymmetric unit. Details of the data collection statistics for hPLK and in complex with MgATP are given in Table 1.
Structure determination of unliganded hPLK
The crystal structure of unliganded hPLK was solved using the Web-tool CaspR, which executes an optimized molecular replacement procedure (Claude et al. 2004). The hPLK1 sequence and the search models of shPLK (PDB code 1LHP; 87% sequence identity to human PL kinase) and ePLK1 (PDB code 2DDM; 24% identity) were first used to derive multiple structure-sequence alignments with the program T-Coffee (Notredame et al. 2000), followed by homology model building with the program MODELLER (Sali and Blundell 1993). Fifteen models were generated with different spatial conformations, and each was automatically screened in search of a molecular replacement solution using AMoRe (Navaza 1994), followed by refinement using CNS (Brunger et al. 1998). Two of the models had the best convergence, with correlation coefficients of about 0.66 and R-factors of 0.40. The next best solution had a correlation coefficient of 0.40 and an R-factor of 0.53.
The model from the converged solution was used for subsequent structure refinement, by means of CNS, with a bulk solvent correction. The starting model was subjected to rigid body, positional, and simulated annealing refinements with all 2.0 Å crystallographic data to Rwork/Rfree of 33.5% / 38.5%. Some residues were omitted due to weak densities. The model was subsequently adjusted, followed by further cycles of refinements and addition of Na+, PO42−, MPD, and water molecules. The refinement statistics are summarized in Table 1.
Structure determination of hPLK in complex with MgATP
The partially refined hPLK structure without solvent molecules and metal ions was used as the starting model to refine against the diffraction data from the cocrystals of hPLK and ATP. Initial rigid body, positional, and simulated annealing refinements at 2.2 Å resolution to Rwork/Rfree of 26.2% / 32.2% clearly showed ATP, Mg2+ and Na+ bound at the active site of the complex, which were modeled in the structure. Statistics for the final model are listed in Table 1.
Structure determination of hPLK crystallized in the presence of PL or AMP–PCP/PL
The fully refined hPLK structure without solvent molecules and metal ions was used to refine against the 2.4 Å and 2.3 Å resolution data from hPLK crystallized in the presence of PL and AMP–PCP/PL, respectively. The difference maps of the partially refined structures of the two crystals did not show bound AMP–PCP and/or PL. Only an MPD molecule was observed bound to the B6 vitamer binding site in both crystals. Further refinements of these two structures showed virtually no significant difference from the unliganded hPLK structure.
Data deposition
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank with the accession codes 2YXT and 2YXU for the unliganded and MgATP-bound hPLK, respectively.