Targeted High-throughput Sequencing for Hematological Malignancies: A GBMHM Survey of Practice and Cost Evaluation in France
Supplemental digital content is available for this article.
Graphical Abstract
Abstract
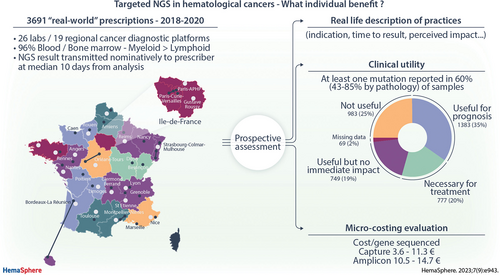
The objective of this study was to assess the clinical impact and financial costs of next-generation sequencing (NGS) in 5 categories of pediatric and adult hematological cancers. NGS prescriptions were prospectively collected from 26 laboratories, with varied technical and reporting practice (all or only significant targets). Impact was defined by the identification of (1) an actionable mutation, (2) a mutation with prognostic and/or theranostic value, and/or (3) a mutation allowing nosological refinement, reported by local investigators. A microcosting study was undertaken in 4 laboratories, identifying the types and volumes of resources required for each procedural step. Individual index prescriptions for 3961 patients were available for impact analysis on the management of myeloid disorders (two thirds) and, mainly mature B, lymphoid disorders (one third). NGS results were considered to impact the management for 73.4% of prescriptions: useful for evaluation of prognostic risk in 34.9% and necessary for treatment adaptation (actionable) in 19.6%, but having no immediate individual therapeutic impact in 18.9%. The average overall cost per sample was 191 € for the restricted mature lymphoid amplicon panel. Capture panel costs varied from 369 € to 513 €. Unit costs varied from 0.5 € to 5.7 € per kb sequenced, from 3.6 € to 11.3 € per target gene/hot-spot sequenced and from 4.3 € to 73.8 € per target gene/hot-spot reported. Comparable costs for the Amplicon panels were 5–8 € per kb and 10.5–14.7 € per target gene/hot-spot sequenced and reported, demonstrating comparable costs with greater informativity/flexibility for capture strategies. Sustainable funding of precision medicine requires a transparent discussion of its impact on care pathways and its financial aspects.
INTRODUCTION
Molecular testing has become an indispensable element in hematological cancer evaluation, as personalized diagnostics are increasingly allowing individualized therapy. Providing appropriate, reproducible, and optimized diagnostics to all patients with cancer is one of the priorities of the European Beating Cancer Plan and its national equivalents. This requires scientific health technology assessment (HTA) and multistakeholder concertation. The French Ministry of Health, through the national cancer institute (Institut National du Cancer [INCa]) has progressively structured cancer care since its creation in 2005 by setting up Multidisciplinary Tumor Boards or MDTs; increasing access to clinical trials in general and in phase I/II reference centers; forming a national network of 28 hospital molecular genetics cancer platforms and creating an HTA funding mechanism for assessment of innovative technologies (Programme de recherche medico-économique; [PRME]). Acronyms are defined in Suppl. Table S1.
To ensure access to innovative in-vitro diagnostics (IVDs) before their inscription on the national biological laboratory tests and procedures schedule (NABM), the French Ministry of Health set up in 2015 a conditional coverage mechanism for a referenced list of innovations not yet included in the national nomenclature (RIHN). Access is free of charge for patients at the point of care, costs being borne by hospitals that in turn receive payment from the ministry. RIHN are given a temporary tariff while the clinical efficacy and economic utility of the tests are evaluated in order to prepare their evaluation by the French National Competent Authority (CA) for Health (HAS) for inscription on the national NABM reimbursement schedule. A significant proportion of RIHN-referenced innovative diagnostic acts corresponds to somatic genetic abnormalities in cancer. Twelve somatic genetic tests involving hematological cancers have received temporary authorization, including 3 proposing reimbursement for targeted next-generation sequencing (NGS) on the basis of the number of kilobases (kb) sequenced (N452 for <20 kb; N453 for 20–100 kb; and N454 for over 100 kb). Some of these molecular analyses underwent economic evaluation in consecutive health economic research programs, in particular the RuBIH1 (2004-6, RéseaU de Biologie Innovatrice en oncoHématologie) network for hematological cancers for polymerase chain reaction (PCR) diagnostics1.,2. and the RuBIH2 assessment of targeted NGS diagnostics, which is the object of this article.
Both RuBIH programs have been undertaken with the French association of molecular biologists for hematological malignancies (Groupe des Biologistes Moléculaires des Hémopathies Malignes [GBMHM]), a nonprofit network, which organizes continuing medical education, concerted actions, and quality assessment3. for molecular diagnostics of hematological cancers. During the RuBIH1 program, the clinical and diagnostic hematology community, under the aegis of the French Hematology Society (SFH), established guidelines for appropriate diagnostic prescriptions2. and undertook HTA assessment of reverse transcription-PCR quantification of fusion transcripts such as BCR-ABL,4. DNA point mutation quantification of single targets such as JAK2 V617F and immunogenetic detection of lymphoid clonality.
The RuBIH2 PRME program (2017–2022; NI6028HLJ) evaluated the place of NGS in the diagnostic evaluation of 5 categories of hematological cancers in adults and children: myelodysplastic neoplasms (MDS); myeloproliferative neoplasms (MPN); acute myeloid leukemia (AML); acute lymphoblastic leukemia (ALL) and lymphoma and mature lymphoproliferative disorders (LPD), including chronic lymphocytic leukemia (CLL). Three approaches were combined: (1) a census of routine diagnostic NGS activity; (2) cost evaluation using a microcosting methodology; and (3) revision by the GBMHM and the SFH of diagnostic prescribing recommendations with regard to guidelines for NGS prescription. The latter anticipated a 2022 HAS request for consensus national NGS panel indications and content for all cancers from clinical and diagnostic cooperative groups and scientific societies. The present report describes these approaches, thus contributing to a macroeconomic evaluation of the structural and budgetary requirement for optimal, equal access to NGS diagnostics in a country such as France, with potential for wider extrapolation.
MATERIALS AND METHODS
Targeted NGS panels
Diagnostic NGS strategies vary significantly between centers. Some laboratories use a number of specific panels whereas others used composite panels, reporting on only a subset of the genes tested, according to the type of hematological malignancy. Details of all panels used during this period (2018–2022) are not provided because their precise content evolved during the impact evaluation period, particularly for capture panels. Some laboratories choose to transmit information to prescribers on all abnormalities considered to be significant in all targeted genes/hot-spots, while others only report information relevant to the disease category, prescription request, and/or protocol agreements regarding actionable targets, while maintaining the possibility of more extensive reporting, when necessary.
Group of molecular biologists for hematological malignancies
The GBMHM was heavily inspired by the European initiatives such as the European Society for Laboratory Hematology-Oncology (ESLHO), https://eslho.org and the European LeukemiaNet, https://www.leukemia-net.org/home/. The GBMHM technological platform is accredited as a provider of external quality assessment (EQA) by the HAS CA. Combined education and bi-annual EQA by the onco-hematology diagnostic community have allowed collective, nationwide, improvement in standardization and harmonized reporting of both CE-marked and, predominantly, in-house IVD testing.3.
Impact analysis
To describe the expected impact of NGS molecular diagnosis on clinical decisions and care circuits, a prospective, one-arm multicentric observational study was conducted. Participating centers are all GBMHM members and each undertook to report a predefined number of consecutive NGS prescriptions, depending on their anticipated activity during the evaluation period. There was no a priori sample size calculation for this observational study.
Only clinical NGS, defined as prescribed tests that led to an individual result transmitted to the prescriber and recorded in the patient file with an identifiable time delay between prescription and reporting, was to be included. Research activity, leading uniquely to the deposition of results in clinical trial databases but not communicated to prescribers, was not collected.
Only an expected impact on the decision and care circuit could be measured in the absence of record linkage that would allow full individual patient follow-up. It was, therefore, assumed that the NGS result would be impactful, based on the data collected by the platforms if (1) an actionable mutation was identified, (2) a mutation (or the absence of it) had a prognostic and/or theranostic value, and/or (3) a mutation allowing nosological precision was identified.
The main data collected were as follows: pathology, type of panel, date of each step of the process, category of prescription (diagnostic, prognostic, theranostic, or combinations thereof), and then the category of clinical decision taken following analysis of the NGS test. Only the gender and age of the patients were collected, without any other personal characteristics.
Participating NGS platforms were requested to provide information on all NGS abnormalities considered to be significant in all targeted genes/hot-spots, disease category, prescription request, and/or protocol agreements regarding actionable targets. They were also asked to transmit data on all clinical prescriptions, whether from within the hospital group or following external prescription. Practical difficulties in obtaining this information, particularly for external prescriptions, led to under-reporting of this category, with 4 laboratories only reporting internal prescriptions.
An online questionnaire to capture NGS prescription data was implemented using the clinical data management system CleanWeb (Telemedicine Technologies, Boulogne-Billancourt, France). NGS prescription data were entered by site investigators, predominantly the molecular diagnostic staff, between October 2018 and November 2020. The origin of the data was the NGS prescription itself, minutes of MDT meetings or hospital information systems. The data are stored by Telemedicine on a secure internet hosting platform conforming to ISO27001 norms. Data analysis on the anonymous data was carried out using the open-source integrated software environment R, and Microsoft Office Professional Plus Excel version 16. The study was approved by the Assistance Publique-Hôpitaux de Paris (AP-HP) ethics committee CERAPHP.5 and the promoter (AP-HP) signed a commitment of compliance with the MR003 reference methodology of the French data protection agency (CNIL) on November 30, 2017.
Cost study
A microcosting study was conducted between 2018 and 2022 at 4 molecular diagnostic platforms in France, where NGS is performed for 5 classes of hematological malignancies. The study was conducted from the health care provider perspective.5.
After identifying types and volumes of resources required for each step of the analytic process, health economists conducted observations in situ. Data-collection spreadsheets were designed based on the workflow provided by the platforms. Personnel time, equipment and consumables (disposables and reagents), and corresponding unit costs were collected to generate an overall average cost. The calculation of production costs was for the full process up to delivering the results to the clinician with the exception of the preanalytic stage of sample collection and DNA extraction. The cost, including human resources, of archiving the complete NGS data set (eg, FASTq files) for 10 years and algorithmic pipeline filtering of data was estimated at 2 centers (No 1 and 2), including by the central bioinformatic (MOABI) platform for the 8 AP-HP laboratories. For 2 platforms (No 6 and 19), the human resources required for data archiving and pipeline filtering were included in the total cost of NGS and were not individualized.
The estimate of personnel costs was based on the time spent on each task and valued using the average AP-HP salary costs for each category of staff, based on the French annual contractual working time of 1607 hours per year. Consumable costs were based on actual consumption and real unit prices, which include negotiated reductions of catalogue prices. Following standard practice, investment costs of the materials were depreciated over 7 years (for large apparatus) or 2 years (for small apparatus)6. using a 2.5% discount rate to reflect opportunity costs of equipment investment.5. Hardware maintenance was calculated as 8% of the unit price of purchase. While no reruns due to problems with sampling, DNA quality, or unexpected results were noted during in situ observations, the possibility of errors requiring reruns for a specific sample was accounted for in the base case by applying a 5% markup to human resource and consumable costs to account for errors based on the expert advice from the study investigators. A deterministic sensitivity analysis was carried out to evaluate costs for a 1% and 10% markup. When it was not possible to observe a step in the process, such as analysis of the results in MDT meetings, the medical biologists were asked to estimate the time involved in such data interpretation.
Average total costs were estimated as a function of annual sample throughput between 100 and 2000 samples for a molecular diagnostic platform and were inflated by 20% to take into consideration overheads such as power, telephone, cleaning, water, logistics, and hospital administration. The estimates included 2 options regarding the allocation of equipment. While the first option assumed a theoretic point of view that the equipment was dedicated to the pathology and panel of genes being analyzed, the second one simulated a polyvalent laboratory in which the equipment was shared between different pathologies, panels, or services. For comparison between panels, an annual throughput of 500 samples for each type of NGS analysis was used for the base case analysis of costs. We estimated costs for routine practice and for complex cases as dictated by the national protocols.7. The unit costs collected for materials in 2018 were inflated using the French consumption of care and medical goods annual indices to 2021€ values.8. The cost analyses were performed with Microsoft Office Professional Plus Excel version 16.
RESULTS
Activity and impact analysis
Twenty-seven laboratories provided prospective recording of all clinical prescriptions, in the 5 categories for which they performed targeted NGS. Details of 4356 NGS prescriptions between October 2018 and November 2020 were collected, but 395 prescriptions were excluded because they were doublets (n = 10), did not respect inclusion criteria regarding pathology or date (n = 18), had insufficient annotations (n = 1), or were nonindex (ie, follow-up samples, n = 366). The latter (1–4 samples/patient), including 132 MDS or MPN, 127 acute leukemia, and 107 LPD prescriptions, were not analyzed further. The 3961 index prescriptions from 3961 patients came from 26 laboratories/hospitals, corresponding to 19 INCa platforms, because 8 belonged to the single AP-HP platform. All but 1 of the 12 mainland French regions were represented, with 1–3 platforms per region. Thus, 19 of the 28 INCa molecular diagnostic platforms were included in this study.9.
Overall, 2584 of 3961 (65.2%) prescriptions were for myeloid disorders and 34.8% for, mainly mature B, lymphoid disorders (Table 1). Prescriptions were mainly at diagnosis/first line of treatment (69%) and showed a male predominance (58%). Age and gender by pathology (Suppl. Figure S1A-S1D) showed that 34% of the prescriptions were from patients aged 70 years or more and only 5% under 20 years. The vast majority of samples were from peripheral blood (PB) or bone marrow (BM) (96%), while lymph node or tissue-derived NGS was rare (3%), in contrast to solid tumor diagnostics. Cell-free DNA samples were not included, because they were mainly experimental in 2018–2020. NGS prescribers were essentially clinical hematologists (89%), with only 6% of prescriptions described as coming via laboratory hematologists, hematopathologists or MDTs, and 4% from other medical specialties.
After exclusion of 871 prescriptions from the 4 platforms for whom reported prescription was restricted to local samples, details of the geographic localization of prescriptions, relative to the INCa molecular platform, are illustrated in Figure 1 for the 15 platforms (3090 results) with exhaustive reporting. Of the 3090 NGS analyses, 61% were performed for patients from the same hospital/INCa platform (n = 1874) and 24% from another hospital within the same department or region (n = 745). Only 15% of samples were transmitted from another region (n = 471), mainly to Parisian and regional reference centers. Approximately half (8/15) of INCa platforms with full description of clinical NGS activity reported prescriptions from another region.
| Patient and Prescription Information | n | % |
|---|---|---|
| Age groups (yrs) | ||
| 0–20 | 201 | 5.1% |
| 20–40 | 344 | 8.7% |
| 40–60 | 922 | 23.3% |
| 60–80 | 2077 | 52.4% |
| 80–100 | 412 | 10.4% |
| MD | 5 | 0.1% |
| Gender | ||
| Male | 2302 | 58.1% |
| Female | 1658 | 41.9% |
| MD | 1 | 0.0% |
| Pathologies | ||
| MPN | 867 | 21.9% |
| MPN/MDS | 184 | 4.6% |
| MDS | 738 | 18.6% |
| AML | 795 | 20.1% |
| ALL | 246 | 6.2% |
| NHL/mature LPD | 1131 | 28.6% |
| Line of treatment | ||
| Diagnostic/First line | 2745 | 69.3% |
| Second line or more | 1098 | 27.7% |
| Post-allograft | 69 | 1.7% |
| Other/unknown | 49 | 1.2% |
| Type of sample | ||
| Blood | 2018 | 50.9% |
| Bone marrow | 1781 | 45.0% |
| Lymph node or other tissue | 126 | 3.2% |
| Othera | 31 | 0.8% |
| MD | 5 | 0.1% |
| Prescriber | 3961 | |
| Clinical hematologist | 3527 | 89.0% |
| Other medical specialtiesb | 152 | 3.8% |
| Diagnostic hematologist | 127 | 3.2% |
| Multidisciplinary tumour board | 89 | 2.2% |
| Pathologist | 59 | 1.5% |
| MD | 7 | 0.2% |
- a Includes 8 CSF, 10 Pleural/pericardiac effusions, 8 cytogenetic pellets and 5 DNAs of unknown origin.
- b Other medical specialties included 7 from medical oncology.
- ALL = acute lymphoblastic leukemia; AML = acute myeloid leukemia; LPD = lymphoproliferative disorders; MD = missing data; MDS = myelodysplastic neoplasms; MPN = myeloproliferative neoplasms; NGS = next-generation sequencing.
An NGS result was generated for 3945 of 3961 prescriptions, corresponding to a failure rate of 0.4%, primarily due to insufficient infiltration or failed extraction/DNA quality, mainly from LPDs (12/16 samples) and non-PB/BM samples (9/157, 5.7% compared with 7/3799, 0.18% of PB/BM; P < 0.00001). Sampling was performed on (n = 3227) or after (n = 115; 50% within 1 week) the day of NGS prescription for 87.6% of the 3813 liquid samples with available data, compared with 21.4% (27/126) of tissue samples, in keeping with a case by case approach by hematopathologists for lymphoma NGS prescription, compared with more frequent up-front NGS in liquid tumor diagnostics. For pre-existing liquid and tissue samples (576, including 6 of undefined origin), the median delay between sampling and NGS prescription was 16 days (range, 1–353), with no significant difference between liquid (median 14 days) and tissue samples (median 16 days). Results were validated and transmitted to the clinicians at a median of 10 days after performing NGS (range, 1–126 days; mean 13 days).
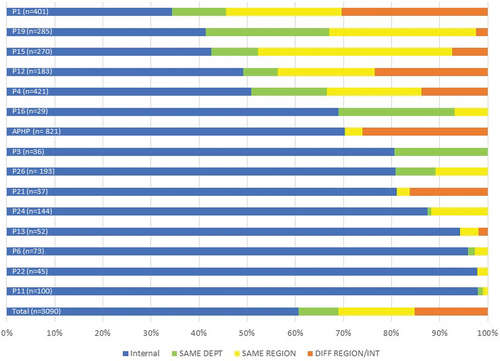
Origin of real-world targeted NGS prescription in hematological cancers. Internal prescriptions (same health institution) are indicated in blue, intradepartment in green, intraregional in yellow and interregional prescriptions in orange. n = total number of reported index prescriptions; NGS = next-generation sequencing.
Detection of at least 1 clinically significant mutation was reported for 60% of the 3945 NGS results transmitted. The highest proportion was identified in AML (85%), interface MPN/MDS (82%), and ALL (77%) (Figure 2A) and the lowest in mature LPDs (43%) and MPN (47%).
Details of the impact of NGS are shown in Figure 3A and 3B. NGS platforms first classified pretest prescribing indications as follows: diagnostic, prognostic, theranostic, or combinations thereof, for 81% of samples, with the remaining 19% also integrating notions of remission monitoring, evolution, response to treatment and relapse (data not shown). NGS results were considered to impact patient management for 73.4% of the prescriptions (Figure 3A), most commonly following MDT discussions and/or protocol-driven treatment modification (Figure 3B). Impact was subclassified as useful for evaluation of prognostic risk in 34.9% (n = 1383) and necessary for treatment adaptation (actionable) in 19.6% (n = 777), but having no immediate individual therapeutic impact in 18.9% (n = 749), with details missing for 1.7% (n = 69) (Figure 3A). The distribution of perceived impact varied between disease subtypes (Figure 2B), the absence of impact being lowest in NHL/LPDs and MPN/MDS. As such, the incidence of detected mutations does not correlate with the perceived impact on patient management, as seen in LNH/LPDs. Treatment advice was specified for 1330 (33.9%) prescriptions and depended on the clinical situation, as detailed in Figure 3B. Comparison of the perceived expected (preanalysis) and actual (postanalysis) impact of NGS for the 2421 prescriptions with available information showed no-change in 59% of analyses and at most only minor modifications in 96% (Suppl. Table S2).
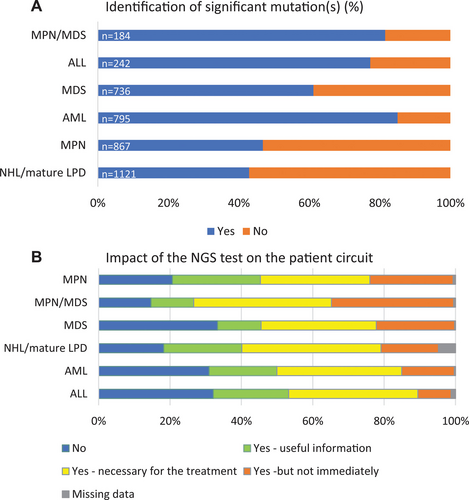
Significant mutations and NGS impact by disease subtype. (A) Significant mutations by disease subtype (n = 3945). Significant mutations are indicated in blue, absence thereof in orange. (B) Impact of the NGS test on the patient pathway by disease subtype (n = 3961). n = total number of reported index prescriptions; NGS = next-generation sequencing.
Cost study
Costing analyses for NGS were performed in 4 independent INCA platforms (N° 1, 2, 6, 19). NGS approaches were all targeted custom panels (amplicon or capture) and included the following: a limited first-line mature lymphoid amplicon approach (LPD/CLL N°2); a pan-hemato-oncology (Hem-Onc) capture approach (N° 19); a single disease, highly protocol-driven, approach typical for a reference center (T-ALL N°2), and 4 myeloid panel approaches, with Platform N°1 comparing manual and automated capture approaches. Unit costs as a function of annual activity are shown in Figure 4 and NGS panel details and breakdown and comparison of costs for an annual activity of 500 tests per year are shown in Table 2.
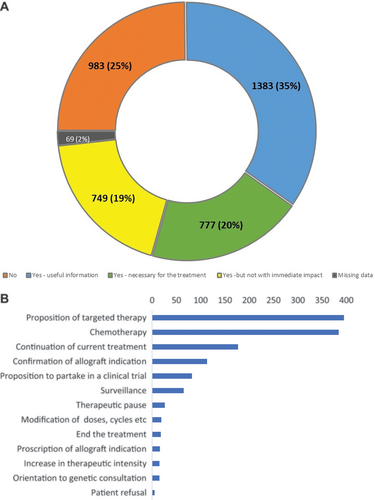
Clinical impact of NGS. (A) Immediate impact of the NGS results (n = 3961). (B) Treatment advice (n = 1330). (B) Treatment advice (n = 1330). NGS = next-generation sequencing.
Comparison of overall costs per result showed that unit costs were higher when using dedicated equipment, particularly for lower annual throughputs (Figure 4A). With shared equipment (occupation estimated at 20%) and uniform personnel costs (AP-HP rates), unit costs at lower levels of activity were dependent on the annual throughput for each type of NGS panel.
| Center 2 | Center 19 | Center 6 | Center 1 | |||||
|---|---|---|---|---|---|---|---|---|
| LPD/CLL | T-ALL Protocol | T-ALL Cognitive | Pan-Hem-onc | Pan-Myeloid | AML | AML | Robot AML/MDS | |
| Panel information | ||||||||
| Method | Amplicon/ Qiaseq |
Capture/ Illumina |
Capture/ Illumina |
Capture/ Haloplex |
Capture/ Haloplex |
Amplicon/ Ionfragment |
Capture/Haloplex | Capture/ Agilent |
| Kilobase (NABM nomenclature) | 24 (N453) | 780 (N454) | 780 (N454) | 324 (N454) | 427 (N454) | 76 (N453) | 76 (N453) | 100 (N454) |
| No. of NGS target genes/hot-spots sequence | 13 | 102 | 102 | 110 | 77 | 36 | 44 | 38 |
| No. of target genes reported on to prescribers | 13 | 5 | 102 | 44 | 64 | 36 | 44 | 38 |
| Microcosting information | ||||||||
| Samples/run at time of evaluation | 19 | 33 | 33 | 24 | 48 | 18 | 14 | 8 |
| Date of microcosting | 2018a | 2018a | 2018a | 2018a | 2018a | 2018a | 2018a | 2022 |
| Total cost of equipment | 106,236 € | 296,908 € | 296,908 € | 348,369 € | 288,393 € | 431,647 € | 561,115 € | 154,000 € |
| Average costs of the hypothesis for 500 samples/yr for shared platforms and 5% rerun rate | ||||||||
| Cost of major equipment/sample maintenance included | 22.0 € | 39.7 € | 39.7 € | 41.5 € | 35.2 € | 60.4 € | 72.7 € | 25.9 € |
| Cost of consumables/sample | 54.2 € | 197.8 € | 197.8 € | 195.4 € | 241.7 € | 174.3 € | 209.3 € | 275.8 € |
| Cost of human resources/sample | 75.2 € | 62.6 € | 124.4 € | 92.5 € | 151.9 € | 74.4 € | 74.1 € | 48.5 € |
| Cost of bioinformatics/sampleb | 7.9 € | 8.8 € | 8.8 € | 0.0 € | 0.0 € | 8.8 € | 8.8 € | 8.8 € |
| Structural costs/sample | 31.4 € | 59.9 € | 72.3 € | 63.7 € | 83.9 € | 60.9 € | 69.5 € | 70.9 € |
| Cost/sample | 190.7 € | 368.8 € | 443.0 € | 393.1 € | 512.6 € | 378.8 € | 434.4 € | 429.9 € |
| Cost/Kb sequenced | 7.9 € | 0.5 € | 0.6 € | 1.2 € | 1.2 € | 5.0 € | 5.7 € | 4.3 € |
| Cost/target sequenced | 14.7 € | 3.6 € | 4.3 € | 3.6 € | 6.7 € | 10.5 € | 9.9 € | 11.3 € |
| Cost/target reported | 14.7 € | 73.8 € | 4.3 € | 8.9 € | 8.0 € | 10.5 € | 9.9 € | 11.3 € |
| Average costs of the hypothesis for 500 samples/yr for shared platforms | ||||||||
| Cost/sample (1% rerun rate) | 184.8 € | 356.9 € | 428.3 € | 379.9 € | 494.7 € | 367.4 € | 421.4 € | 415.2 € |
| Cost/sample (10% rerun rate) | 198.1 € | 383.7 € | 461.4 € | 409.5 € | 535.1 € | 393.0 € | 450.6 € | 448.5 € |
- Total costs/sample for 500 samples/yr on a shared platform with a 5% rerun rate are shown in bold.
- a Updated in 2022.
- b Per sample bioinformatic costs include data storage and algorithmic pipeline analysis before validation, evaluated in centers 1 and 2. Equivalent costs were not individualized by centers 6 and 19.
- ALL = acute lymphoblastic leukemia; AML = acute myeloid leukemia; CLL = chronic lymphocytic leukemia; LPD = lymphoproliferative disorders; MDS = myelodysplastic neoplasms; NGS = next-generation sequencing.
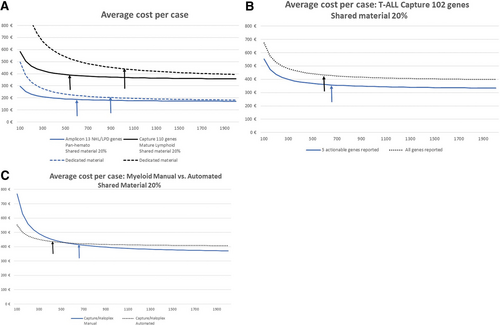
Average cost in Euros per case as a function of annual activity. Vertical arrows indicate activity thresholds for which a change in ±50 annual NGS analyses did not result in a change in unit cost of >±1%. (A) Average cost per case. (B) Average cost per case: T-ALL capture 102 genes shared material 20%. (C) Average cost per case: myeloid manual vs automated shared material. ALL = acute lymphoblastic leukemia; NGS = next-generation sequencing.
The average overall cost per sample was, as expected, much lower for the restricted, first-line mature lymphoid LPD/CLL amplicon panel (191€; 5% rerun rate) compared with all others (Table 2), when the cost of capture panels varied from 368€ to 513€. Unit costs varied more as a function of the number of targets reported (5–102) than kb (24–780) sequenced. Unit costs varied from 0.5 to 7.9€ per kb sequenced, from 3.6€ to 14.7€ per target gene/hot-spot sequenced, and from 4.3 to 73.8€ per sample for target gene/hot-spot reported. Extremes are represented by the 2 formats used to report data from the T-ALL panel. Reporting of only the 5 actionable T-ALL genes, compared with reporting relevant mutations for all 102 genes in relapse/resistant or complex cases (Figure 4B) led to a doubling of human resource unit costs (essentially medical) from 63€ to 124€ and an increase in overall costs of 20% (369€ to 443€). Based on the local estimation that 75% of samples require reporting on only 5 targets and 25% require full analysis, the average unit cost would be 387€ per sample. This was similar to the unit cost of 393€ using the pan-Hem-Onc panel (platform N° 19), reporting on up to 44 genes, with the actual number reported depending on the prescription and clinical situation. Comparison of manual versus automated costs for a myeloid capture panel on platform N°1 (Figure 4C) showed that overall unit costs were slightly higher following automation at higher annual throughput, with a significant increase in the cost of consumables, partially offset by a decrease in human resources, which can therefore be diverted elsewhere, and equipment costs. This was, however, reversed at lower levels of activity. This can be explained by the low number of samples processed in a single run, which results in higher sequencing costs but makes it possible to obtain a result within 48 hours of receiving the sample. Automation also allows for more robust and reproducible NGS workflows.
The cost of data storage (eg, FASTQ formatted files) and algorithmic pipeline filtering before validation was not the focus of this cost study but was estimated by centers 1 and 2 and is shown in Table 2 and Suppl. Table S3. Centers 6 and 19 included these costs in the overall cost estimate for the NGS analysis.
The activity thresholds for which a change in ±50 annual NGS analyses did not result in a change in unit costs of >±1% (flat part of the cost/activity curve) ranged between 550 and 700 analyses per year, except for the automated myeloid panel (450/year.).
Taken together, these costing analyses demonstrate that minimal annual activity levels of over 500 samples are acceptable from an economic point of view, but that automation may allow lower activity levels while maintaining resource efficiency. They also show that a single, uniform reimbursement policy for targeted NGS is not appropriate, because it is unlikely to incite economic optimization.
DISCUSSION
This observational, real-world study of 3961 index targeted NGS prescriptions in 5 categories of hematological cancers found that the analyses provided clinically relevant information for 75% of the patients. Prescriptions were essentially PB or BM (3% tissue) and were mainly (65%) for myeloid disorders. Approximately two thirds of INCa platforms (19/28) participated in this study, but with variable contributions and representation of reference laboratories specializing in a limited number of disease categories. Bearing in mind these limitations, reported prescriptions (Table 1) corresponded to 7.8% of MDS, 8.5% of MPN and MDS/MPN, 12% of AML, 13.7% of ALL but only 3.2% of NHL/MLPD cases, based on the annual incidences reported in Table 3. As such, <20% of all hematological cancer categories benefit from routine NGS analysis, with the highest coverage in AL and the lowest in NHL/MLPD. The latter is likely to be particularly true for NHL, based on the second-line strategies for tissue-based diagnostics. ALL, AML, and interface MPN/MDS were the conditions where NGS was most likely to detect mutations. Of note, the absence of certain mutations can also be actionable, for instance, the absence of NOTCH1/FBXW7 mutations in adult T-ALL defines a poor prognosis group.7. The median turnaround time to reporting was 10 days (mean 13 days). No attempt was made in this study to standardize NGS reporting, but such measures are underway nationally with INCa and at the European level, for example in the CanHeal: https://canheal.eu/ program.
The cost per analysis for an (arbitrary) annual testing volume of 500 samples ranged roughly from 200€ for amplicon to 350–500€ for capture NGS, depending on the number of targets. Although the automated cost analysis was performed 4 years after the, albeit updated, manual analysis and no comparison of technical performance was done, it did demonstrate the shifting budgetary impact (from personnel to consumables) and lower unit costs for lower levels of activity, which automation can provide. From an HTA point of view, the activity threshold that renders unit costs relatively independent of annual activity is probably around 700 samples per year with current equipment and processes. Recent NGS cost evaluations report highly variable unit costs10. thus complicating reimbursement considerations.
Bioinformatics costs was not the focus of this analysis but 2 centers performing in-house, relatively high throughput, with central facilities, both estimated this to be ≈9€ per sample including the long-term data storage, before taking into consideration infrastructure and amenities such as electricity or pipeline development. This represents between 2% and 5% of the total cost of the NGS analysis. For comparison, an Australian study estimated, in 2020, the cost of long-term data storage of the whole genome to be 2.6% of the total sequencing cost.11. Such costs are likely to be significantly higher if out-sourced. The bioinformatic costs cited here were calculated by public sector bioinformatic teams and did not include the cost of initial algorithm design or intellectual property (IP) costs. The diagnostic community should be aware of the significant differences in unit costs, due indirectly or directly to outsourcing and the added costs of IP in the manufacturing sector. These considerations will also apply to the cost of CE-marked NGS assays, particularly after application of IVD Regulation 2017/746.12. It is premature to estimate the economic impact of this European legislation, because it will impact many aspects of diagnostic practice, with probable increases in unit costs, potentially partially offset by increasing centralization.
This real-life study complements evidence from clinical trials and meta-analyses.13. It also has the limitations of a study using data collected at different time points from platforms that volunteered. We attempted to reduce bias by requesting that consecutive patients be included. Impact analysis particularly for identifying care pathway prescriptions from outside the platform’s hospital was, however, complicated by the regulatory authorizations required. This currently constitutes a major obstacle for any real-world impact study in precision medicine that cannot easily link sequencing data to medical outcome and trial registries. We recommend as an initial step that all consent forms be adapted to allow linkage of sequencing data to claims data and electronic medical records.
France has ensured financial equity in access to precision medicine through its publicly funded sequencing platforms. This study shows that 85% of NGS prescriptions are performed at the local/regional level and 15% in national reference centers. The ongoing implementation of a network of reference laboratories, in concertation with the INCa and HAS, will progressively ensure equity in access. Given their relative rarity, and participation in both clinical trials and HTA PRME programs, this process is further advanced for hematological cancers than solid tumors. Currently, of the 864 hospitals, which have an authorization to treat cancer (all tumors) patients, only 126 are tertiary care or comprehensive cancer centers and both treat about 36% of all cancer patients in France.14.
Sustainable funding of precision medicine requires a transparent discussion of its financial aspects, with involvement of relevant medical specialists. Value-based payment for precision diagnostics is complicated because their real value can only be measured with the subsequent care-management and is therefore highly dependent on the cost effectiveness of the treatments applied. The fallback position is to use production costs as a proxy for prices and reimbursement. This means that up-to-date cost studies need to be conducted to help public payers reduce the financial benefit that some laboratories derive from public reimbursement. The type of panel chosen, with its corresponding reimbursement, should be dictated by the category of samples received and the local front-line versus regional versus national reference nature of the platform. For clinical practice, a reimbursement policy should incite maximal provision of actionable information, which must be preceded by rigorous pre and postmarketing surveillance, including in real-world situations. Of note, the value of NGS is not limited to decisions on actionable treatment choice because, for example, it also allows minimal residual disease assessment. The costing study presented here suggests that targeted NGS reimbursement should be based on the number of evaluated and/or reported targets, rather than the number of kilobases sequenced. We would recommend 2 categories of panel (with appropriate reimbursement): one with <20 targets, amenable to amplicon approaches, and one with 20 or more targets, for which capture NGS is likely to be more appropriate. Cost comparisons showed, however, that from an economic point of view unit costs of capture panels compare favorably with amplicon panels, while providing greater flexibility and universality.
Because payment is activity-based, guidelines are needed to ensure that only the appropriate analyses are performed and that the necessary expertise is available for optimal interpretation, although this does not necessarily need to be local. Achieving economically optimized activity thresholds also implies that NGS analyses either need to be heavily centralized (eg, platform 2, T-ALL), pan-Hem-Onc (eg, platform 19), or an appropriate combination of these, based on the local case-loads and national agreements regarding local versus reference center activity. It is also important to appropriately combine purely clinical and academic use of NGS panels.
NGS guidelines will be based on the evidence from clinical trials but can also be informed by real-world evidence on the clinical actionability and actual usefulness to change the patients’ journeys, as evaluated here. The recommendations presented here are not intended to constitute guidelines, but more to provide an overall idea of consensus clinical practices in France. They are complementary to recent international recommendations for hematological malignancies15.,16. and should contribute to the calculation of expected national or European requirements for NGS oncogenetics in hematological cancers from the health care provider perspective and for comparison with NGS activity reported by the INCa/HAS or other competent authority platforms.
National consensus for NGS prescribing
The GBMHM published consensus NGS panels for lymphoid cancers in 2019.17. At the request of the HAS, these were complemented in 2022 by recommendations on indications for analysis in routine clinical practice, after concertation with the relevant national clinical cooperative groups, under the aegis of the SFH. These recommendations included NGS indications for immunogenetics (IG/TR) and cell-free/circulating DNA, neither of which are addressed here.
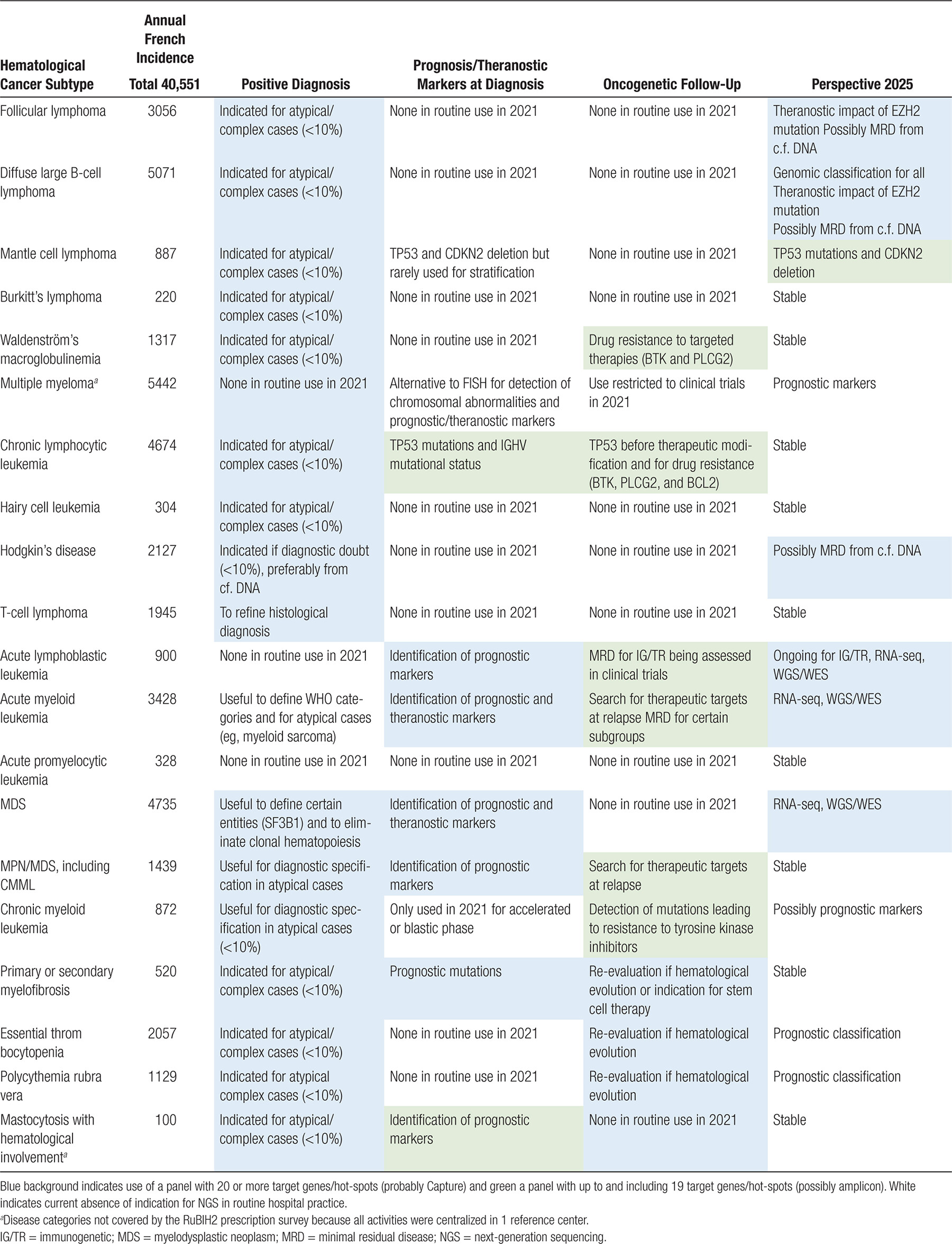
Hematological cancers were divided into 20 categories (Table 3), including 11 lymphoid and 9 myeloid, with an annual incidence ranging from 100 (mastocytosis with hematological involvement) to 5442 (myeloma).7. With over 40,000 new cases of hematological cancer reported each year in France (population 67 million inhabitants in 2018/2019), NGS was recommended for at least 10% of cases in virtually all disease categories, but also for identification of prognostic markers at diagnosis in all cases of CLL, ALL, AML, MDS, MPN/MDS, and myelofibrosis. The latter group corresponded to a total of 15,796 new cases per year. Multiple myeloma was analyzed in a single national reference center, essentially within clinical trials, and, as such, was not detailed, despite its frequency, but was here considered to require NGS in ≈10% of cases at diagnosis. Combining these indications, it would be expected that at least 18,300 NGS oncogenetic analyses (15,800 plus 10% of the remaining 24,800) would be prescribed each year in France for hematological cancers. This is an underestimate of the total NGS activity reported each year, because it did not take into account analysis at (suspected) therapeutic resistance/evolution/relapse, measurable/minimal residual disease monitoring or IG/TR analyses.
In keeping with the cost analyses and types of targeted panels developed by diagnostic laboratories, recommendations for panel use have been divided into panels targeting ≤20 or more genes/hot-spots (Table 3). Individual panel use is up to local choice for custom in-house IVD or CE-IVD NGS assays, in turn dependent on local first-line versus reference recruitment.
These national consensus panels, approved by both the clinical and diagnostic hematology-oncology community, should contribute to the identification of appropriate prescribing practice of targeted NGS and its reimbursement.
COLLABORATORS
*GBMHM Group: Mélissa Alame, Centre Hospitalier Universitaire de Montpellier; Fanny Baran-Marzak, Bobigny-APHP Hôpital Avicenne; Marc G. Berger, Centre Hospitalier Universitaire Clermont-Ferrand – site Estaing; Dominique Bories, Créteil-APHP Hôpital Henri Mondor; Aurélie Caye-Eude, Paris-APHP Hôpital Robert Debré; Jean-Michel Cayuela, Paris-APHP Hôpital Saint Louis; Pascale Cornillet-Lefebvre, Centre Hospitalier Universitaire de Reims; François Delhommeau, Sorbonne University, INSERM, Saint-Antoine Research Center, CRSA, AP-HP, Saint-Antoine Hospital, Paris, France; Marie-Hélène Estienne-Felix, Centre Hospitalier Régional Universitaire de Tours - site Bretonneau; Pascaline Etancelin, Centre Henri Becquerel, Rouen; Pascale Flandrin-Gresta, Centre Hospitalier Universitaire de Saint-Étienne; Eric Lippert, Centre Hospitalier Universitaire de Brest; Christophe Marzac, Villejuif-Institut Gustave Roussy; Laurent Miguet, Hôpitaux Universitaires de Strasbourg; Cédric Pastoret, Centre Hospitalier Universitaire de Rennes; Sophie Raynaud, Centre Hospitalier Universitaire de Nice; David Rizzo, Centre Hospitalier Universitaire de Limoges.
ACKNOWLEDGMENTS
The authors thank Alban Lermine and Guillaume Meurice for costing analyses of bioinformatic support by the AP-HP MOABI platform and all those who provided data and/or contributed to the GBMHM guide to NGS prescription:
Sandrine Geoffroy, Centre Hospitalier Universitaire de Lille; Alice Marceau- Renaut, Centre Hospitalier Universitaire de Lille; David Sibon, Paris-APHP Necker Enfants Malades; Patrick Villarese, Paris-APHP Necker Enfants Malades; Pascale Flandrin-Gresta, Centre Hospitalier Universitaire de Saint-Étienne; Stéphanie Dulucq, Centre Hospitalier Universitaire de Bordeaux; Fanny Robbesyn, Centre Hospitalier Universitaire de Bordeaux; Stéphanie Dufrechou, Centre Hospitalier Universitaire de Toulouse; Laetitia Largeaud, Centre Hospitalier Universitaire de Toulouse; Naïs Prade, Centre Hospitalier Universitaire de Toulouse; Annaëlle Beucher, Centre Hospitalier Universitaire Angers; Odile Blanchet, Centre Hospitalier Universitaire Angers; Anne Bouvier, Centre Hospitalier Universitaire Angers; Bruno Cassinat, Paris-APHP Hôpital Saint Louis; Jean-Michel Cayuela, Paris-APHP Hôpital Saint Louis; Michaël Deagaud, Paris-APHP Hopital La Pitié-Salpetriere; Clotilde Bravetti, Paris-APHP Hopital La Pitié-Salpetriere; Emmanuelle Clappier, Paris-APHP Hôpital Saint Louis; Dominique Bories, Créteil-APHP Hôpital Henri Mondor; Ivan Sloma, Créteil-APHP Hôpital Henri Mondor; Sihem Tarfi, Créteil-APHP Hôpital Henri Mondor; Chloé Arfeuille, Paris-APHP Hôpital Robert Debré; Aurélie Caye-Eude, Paris-APHP Hôpital Robert Debré; Pascale Cornillet-Lefebvre, Centre Hospitalier Universitaire de Reims; Valérie Gonçalves, Centre Hospitalier Universitaire de Reims; Angéline Preto, Centre Hospitalier Universitaire de Reims; Marie Laure Boulland, Centre Hospitalier Universitaire de Rennes; Cédric Pastoret, Centre Hospitalier Universitaire de Rennes; Pascaline Etancelin, Centre Henri Becquerel, Rouen; Laurent Miguet, Hôpitaux Universitaires de Strasbourg; Marie-Christine Bené, Centre Hospitalier Universitaire de Nantes; Audrey Ménard, Centre Hospitalier Universitaire de Nantes; Aurelie Chauveau, Centre Hospitalier Universitaire de Brest; Eric Lippert, Centre Hospitalier Universitaire de Brest; Chloé Friedrich, Paris-APHP Hôpital Cochin; Loria Zalmai, Paris-APHP Hôpital Cochin; Fanny Baran-Marzak, Bobigny-APHP Hôpital Avicenne; Sarah Huet, Hospices Civils de Lyon, Hôpital Lyon Sud; François Delhommeau, Sorbonne University, INSERM, Saint-Antoine Research Center, CRSA, AP-HP, Saint-Antoine Hospital, Paris, France; Fabrizia Favale, Sorbonne University, INSERM, Saint-Antoine Research Center, CRSA, AP-HP, Saint-Antoine Hospital, Paris, France; Pierre Hirsch, Sorbonne University, INSERM, Saint-Antoine Research Center, CRSA, AP-HP, Saint-Antoine Hospital, Paris, France; Christophe Marzac, Villejuif-Institut Gustave Roussy; Nathalie Gachard, Centre Hospitalier Universitaire de Limoges; David Rizzo, Centre Hospitalier Universitaire de Limoges; Pascal Turlure, Centre Hospitalier Universitaire de Limoges; Mélissa Alame, Centre Hospitalier Universitaire de Montpellier; Marc G. Berger, Centre Hospitalier Universitaire Clermont-Ferrand – site Estaing; Celine Bourgne, Centre Hospitalier Universitaire Clermont-Ferrand – site Estaing; Benjamin Lebecque, Centre Hospitalier Universitaire Clermont-Ferrand – site Estaing; Thomas Cluzeau, Centre Hospitalier Universitaire de Nice; Sophie Raynaud, Centre Hospitalier Universitaire de Nice; Fabienne Vasquez, Centre Hospitalier Universitaire de Nice; Marie-Hélène Estienne-Felix, Centre Hospitalier Régional Universitaire de Tours - site Bretonneau.
AUTHOR CONTRIBUTIONS
MD, CP, ID-Z, and EM designed the study and wrote the article. PS, OK, DLP, and ASA coordinated the collection and compilation of GBMHM indications for NGS analysis. PS, DLP, MF, SK, and EM provided microcosting data to MD, SC, NM, and ID-Z. PS, OK, DLP, SK, SH, ASA, AB, YLB, ED, and FD participated in data generation and performance of research. MD, EM, CP, SC, ID-Z, and GBMHM members participated in data analysis.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
This work was supported by a grant from the Direction Générale de l’Offre de Soins (DGOS) of the French Ministry of Health (reference number: PRME-K16-014 - RUBIH2).