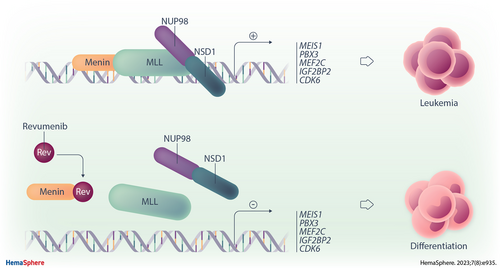
The MLL–Menin Interaction is a Therapeutic Vulnerability in NUP98-rearranged AML
Supplemental digital content is available for this article.
Graphical Abstract
Abstract
Chromosomal translocations involving the NUP98 locus are among the most prevalent rearrangements in pediatric acute myeloid leukemia (AML). AML with NUP98 fusions is characterized by high expression of HOXA and MEIS1 genes and is associated with poor clinical outcome. NUP98 fusion proteins are recruited to their target genes by the mixed lineage leukemia (MLL) complex, which involves a direct interaction between MLL and Menin. Here, we show that therapeutic targeting of the Menin–MLL interaction inhibits the propagation of NUP98-rearrranged AML both ex vivo and in vivo. Treatment of primary AML cells with the Menin inhibitor revumenib (SNDX-5613) impairs proliferation and clonogenicity ex vivo in long-term coculture and drives myeloid differentiation. These phenotypic effects are associated with global gene expression changes in primary AML samples that involve the downregulation of many critical NUP98 fusion protein-target genes, such as MEIS1 and CDK6. In addition, Menin inhibition reduces the expression of both wild-type FLT3 and mutated FLT3-ITD, and in combination with FLT3 inhibitor, suppresses patient-derived NUP98-r AML cells in a synergistic manner. Revumenib treatment blocks leukemic engraftment and prevents leukemia-associated death of immunodeficient mice transplanted with NUP98::NSD1 FLT3-ITD-positive patient-derived AML cells. These results demonstrate that NUP98-rearranged AMLs are highly susceptible to inhibition of the MLL–Menin interaction and suggest the inclusion of AML patients harboring NUP98 fusions into the clinical evaluation of Menin inhibitors.
INTRODUCTION
More than 50% of pediatric and adolescent acute myeloid leukemia (AML) cases are characterized by chromosomal rearrangements that give rise to leukemic fusion genes.1. Translocations of the Nucleoporin 98 (NUP98) gene are among the most common recurrent translocations in pediatric AML, representing 4% of all fusion events. Internal tandem duplications in the Fms-like tyrosine kinase 3 gene (FLT3-ITD) are a common genetic alteration in NUP98-rearranged (NUP98-r) AML (>70%),2. which has been associated with a dismal clinical outcome.3.
The wild-type NUP98 protein contains 2 N-terminal intrinsically disordered regions featuring 38 FG/GLFG repeats, which are separated by a GLEBS motif. This intrinsically disordered FG-rich region is involved in trafficking of RNA molecules through the nuclear membrane and participates in the formation of biomolecular condensates.4. The C-terminal region of wild-type NUP98 contains an RNA-binding domain and an autoproteolytic cleavage site, where the NUP98 precursor protein is cleaved into 90 and 8 kDa peptides.5. In addition, it participates in the regulation of gene expression within biomolecular condensates that were termed GLFG bodies.4.,6. Without exception, all NUP98 fusions join the N-terminus of NUP98 including the FG repeats to the C-terminus of fusion partners, which often contain domains with roles in epigenetic modifications and/or transcriptional control. More than 30 gene partners have been reported for NUP98 in a wider range of AML subtypes including monocytic, megakaryoblastic, and erythroid AMLs, with NUP98::NSD1 and NUP98::KDM5A as the most prevalent.2.,3.,7.,8. The resulting fusion proteins promote leukemia by recruiting various chromatin regulatory proteins, leading to the transcriptional activation of leukemia-associated genes.9.-11.
NUP98-rearranged (NUP98-r) AML is characterized by a distinct gene expression pattern including elevated expression of HOXA and MEIS1 genes, which is similar to that of NPM1-mutated (NPM1-mut) and MLL-r AML, with the exception that HOXB genes are also overexpressed in NUP98-r and NPM1-mut AMLs.3.,12. Recently, we have also demonstrated that NUP98-r AMLs, along with NPM1-mut and MLL-r AMLs, cluster together in principal component analyses.13. In line with this shared transcriptional pattern, NUP98 fusion proteins interact with various chromatin-modifying complexes, including mixed lineage leukemia (MLL) complexes. MLL has been shown to recruit NUP98 fusion proteins to its target genes and is required for the gene expression signature and proliferation of NUP98-r AML cells.14. The interaction between the N-terminal moiety of MLL and its cofactor Menin is critical in MLL-r leukemia, and therapeutic disruption of this association represents a promising targeting strategy that is under current clinical evaluation for this AML type. To date, 3 different Menin inhibitors (SNDX-5613, revumenib; KO0539, ziftomenib; JNJ-75276617) are under clinical investigation with all 3 inhibitors cause dissociation of the Menin–MLL complex. Unlike its 2 sister compounds, revumenib is a CYP3A4 substrate resulting in parallel dose finding studies with and without CYP3A4 inhibitors.15. These phase 1 studies have already shown that inhibiting the MLL–Menin interaction with Menin inhibitors reverses leukemic gene expression and induces differentiation of MLL-r leukemic cells, resulting in clinical remission.15.-19.
The physical interaction of NUP98 fusion proteins with MLL complexes at promoters in combination with the similar leukemia-associated gene signature in NUP98-r, NPM1-mut, and MLL-r AML raises the question whether NUP98-r AML is dependent on the Menin–MLL interaction. Using cell lines and PDX mouse models expressing NUP98::HOXA9, NUP98::JARID1A, and NUP98::NSD1 fusions, Heikamp and colleagues recently demonstrated that interrupting the Menin–MLL interaction disturbs global gene expression and halts leukemic progression in NUP98-r AMLs, but no primary samples were studied.20.
In addition, combined targeting of Menin and secondary lesions such as FLT3-ITDs has not been studied previously in patient-derived NUP98-r AML. In this study, applying an ex vivo coculture approach, we found that primary NUP98-r AMLs are sensitive to the Menin inhibitor revumenib (SNDX-5613) (EHA abstract 2021). Menin inhibition impaired leukemic proliferation and clonogenicity and induced myeloid differentiation in primary patient-derived AML samples that was associated with global gene expression changes. As the vast majority of NUP98-r cases are also FLT3 mutated,2.,21.,22. we combined Menin and FLT3 inhibition, which caused a strong synergistic suppression of the expansion of patient-derived NUP98::NSD1 FLT3-ITD double-positive AML cells ex vivo. Importantly, Menin inhibition completely blocked disease progression by eradicating leukemic blasts in a patient-derived NUP98::NSD1/FLT3-ITD-positive PDX model, suggesting that the NUP98 fusion drives the leukemia. Overall, the results of this study support the clinical evaluation of Menin inhibitors in NUP98-r AML patients.
MATERIALS AND METHODS
Culture of primary AML cells
Primary AML cells were cultured with healthy bone marrow-derived mesenchymal stem cells (MSCs) under serum-free conditions. MSCs were seeded at a density of 7500 cells/cm2 in DMEMlow Glucose plus Glutamax supplemented with 20% fetal bovine serum, 8 ng/mL Fibroblast Growth Factor-beta (FGF2; PeproTech, London, UK) and 100 U/mL penicillin/streptomycin (Gibco BRL, Life Technologies, Breda, The Netherlands), and cultivated at 37°C in 5% CO2 overnight or until reaching 50%–70% confluence. Primary AML cells were thawed and added to the MSC layer at a density of 5 × 105 cells/mL in SFEMII medium supplemented with 100 U/mL penicillin/streptomycin, 10 ng/mL IL-3, 10 ng/mL FLT3 ligand, 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF), 150 ng/mL stem cell factor (SCF), 100 ng/mL thrombopoietin (TPO) (all cytokines were purchased from PeproTech), 750 nM SR1 (Biogeme, Lausanne, Switzerland), and 1.35 µM UM729 (STEMCELL Technologies, Cologne, Germany). Cocultures were incubated at 37°C in 5% CO2 and expanded with readjusting AML cell numbers to 5 × 105 cells/mL every 4 days.
Inhibitor treatment
Primary AML cells were kept in coculture for 4 days, replated on fresh MSCs, and treated with different concentrations of revumenib or 0.1% DMSO (mock). AML cells were replated at a cell concentration of 5 × 105 cells/mL every 4 days on fresh MSCs over a period of 14 days with concomitant inhibitor replenishment (Figure 1A). During this period, untreated leukemic stem cells (CD34+ and CD45RA+) expanded >12-fold with doubling times in the range of 2–3 days. For determining cell viability, the number of viable cells was counted at different time points using 0.4% trypan blue exclusion (Invitrogen, T10282). The ratio of viable cells in DMSO-treated groups versus revumenib-treated groups was plotted to calculate IC50 values (Prism 8, GraphPad). The differentiation status of revumenib-treated or DMSO-treated cells was characterized at day 14 by flow cytometry using CD11b-APC-Cy7 (Biolegend, San Diego, CA), by quantitative polymerase chain reaction (qPCR) using ITGAM (CD11b)-specific and MNDA-specific primers and by Wright-Giemsa-stained cytospin slide preparations. For combinational treatment, serial concentrations of revumenib and gilteritinib were used at a constant 4:1 ratio. The highest concentrations were 2.5 µM and 625 nM for revumenib and gilteritinib, respectively. AML cells were treated with revumenib for 10 days followed by the addition of gilteritinib and cell count on day 13.
In vitro studies and statistical analysis
See the Supplemental Methods for flow cytometry, apoptosis analysis, RNA-sequencing, cytospin analysis, real-time qPCR, clonogenic assay in methylcellulose, western blotting, data quantification, and statistical analysis.
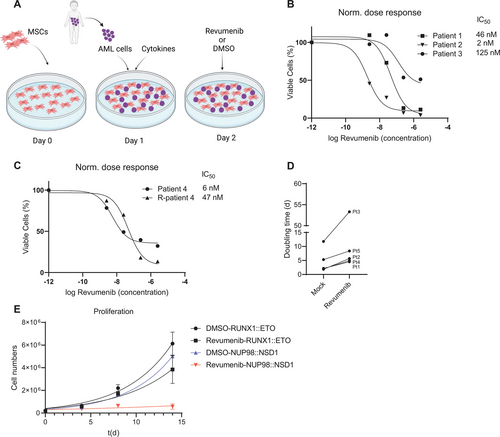
NUP98-rearranged (NUP98-r) AML cell growth is inhibited upon treatment with revumenib. (A) Schematic representation of the human bone marrow-derived MSCs coculture platform used to culture primary AML cells. MSCs serve as feeder layers and support AML cell proliferation. Figure created with BioRender.com. (B and C) Normalized dose-response curve of primary NUP98::NSD1 (FLT3-ITD+) AML cells treated with revumenib for 2 weeks. (D) Doubling time of primary AML cells in coculture treated with DMSO or revumenib (250 nM). (E) Proliferation curves of primary AML cells harboring RUNX1::ETO and NUP98::NSD1 under treatment of revumenib (250 nM). Data were presented as mean (±SD). AML = acute myeloid leukemia; MSCs = mesenchymal stem cells.
AML xenograft model
NSG mice were injected intrafemorally with 106 primary AML cells from patient 1 harboring the NUP98::NSD1 fusion with concomitant FLT3-ITD mutation (Suppl. Table S1). Three months after confirmed engraftment, mice were randomized into 3 groups and were fed on a chow diet of 0.01% revumenib, 0.033% revumenib, and normal chow diet. Peripheral blood was collected at different time points and the fraction of CD45+ CD33+ CD34+ human AML cells was assessed via flow cytometry. Mice were humanly killed upon weight loss exceeding 20%, showing clinical signs of leukemia or at the experimental end point.
RESULTS
Revumenib inhibits growth of primary patient-derived NUP98-r AML cells
To test the sensitivity of NUP98-r AML to the Menin inhibitor, we used transgenic cells ectopically expressing NUP98 fusions and patient-derived primary AML cells containing NUP98 rearrangements. We explored Menin dependency with revumenib, a Menin inhibitor currently under clinical evaluation.23.
We first examined the impact of Menin inhibition on cells transduced with NUP98::NSD1 and NUP98::DDX10 fusion genes.24. Revumenib suppressed cell proliferation of both transduced AML cell lines in a dose-dependent fashion with IC50 values of 56 and 36 nM, respectively (Suppl. Figure S1A-S1C).
Next, we investigated the impact of Menin inhibition on the propagation of patient-derived primary AML cells comprising 5 NUP98::NSD1 and 1 NUP98::TOP1 AML sample. The NUP98::NSD1 samples included a matched sample from the same patient at presentation and relapse (Patient 4). Sequence analysis revealed that all NUP98::NSD1 samples expressed 2 chimeric fusion mRNAs, with the joining of NUP98 exon 12 to NSD1 exon 6 being the predominant transcript. The minor splice variant was the fusion of NUP98 exon 11 and NSD1 exon 6 (Suppl. Figure S1E). Importantly, all NUP98::NSD1 samples carried FLT3-ITD at allelic frequencies >0.4, while the NUP98::TOP1 sample carried a combination of WT1 and CEBPA mutations (Suppl. Table S1). We exposed AML cells to a range of revumenib concentrations between 2.5 and 2500 nM. Coculture on human bone marrow-derived MSCs maintained proliferation of primary AML samples with doubling times ranging from 2 to 10 days (Figure 1D and 1E; Suppl. Figure S1F). Revumenib treatment resulted in a profound dose-dependent suppression of proliferation that was associated with 2-fold increased doubling times and IC50 values ranging from 2 to 120 nM (Figure 1B–1E). In contrast, AML cells expressing the RUNX1::ETO fusion protein were unaffected by revumenib at a concentration of 250 nM (Figure 1E; Suppl. Figure S1D).25. Revumenib inhibited cell proliferation with a 10-day latency, which is consistent with the inhibitory effect of revumenib on NUP98::NSD1 and NUP98::DDX10 transduced cells. This observation is also in agreement with the impact of the Menin inhibitor MI-503 on MLL-rearranged leukemia cells,26.,27. and other inhibitors of epigenetic factors, for example, DOT1L28. and EZH2.29. Menin inhibition resulted in a slight increase in apoptotic cells after 2 weeks, which might be a consequence of myeloid differentiation (Suppl. Figure S2A). Instead, combined histology, qPCR, and flow cytometric analysis revealed a dose-dependent decrease of CD34 and CD45RA and a gain of CD11b, CD14, and MNDA (Figure 2A and 2B; Suppl. Figure S2B-S2F), indicating that revumenib treatment induced myelo-monocytic differentiation. To investigate more comprehensively how Menin inhibition affects differentiation and the distribution of subpopulations of leukemia cells in NUP98::NSD1 (FLT3-ITD+) and NUP98::TOP1 (WT1+, CEBPA+) primary AML cells, we examined the surface expression of CD14, CD33, CD11b, CD38, CD90, CD34, CD117, and CD45RA. The t-stochastic neighbor embedding analysis identified 4 distinct CD34+ cell clusters (Figure 2C). Revumenib treatment led to a strong reduction of all CD34+ cell clusters 1–4 and a corresponding increase in CD34-cell populations. This increase was associated with changes in the distribution of CD14 and CD38 markers within the cell populations. Examination of the clonogenic potential of NUP98::NSD1-harboring cells as a surrogate marker for malignant self-renewal revealed that revumenib reduced colony numbers >50% as compared with DMSO controls, and the remaining colonies had a more dispersed appearance and were smaller (Figure 2D and 2E). These data demonstrate that treatment with revumenib induces myelo-monocytic differentiation in primary NUP98-r cells and suggest that Menin inhibition causes a concomitant loss of self-renewal even in the presence of the FLT3-ITD mutation.
Revumenib induces downregulation of NUP98 fusion target genes including FLT3-ITD
Like MLL-r AMLs, AMLs harboring a NUP98 rearrangement are characterized by high expression of MEIS1 and the HOXA cluster genes. As the expression of these genes was shown to be dependent on a functional MLL–Menin interaction,10. we examined the impact of inhibiting this interaction on the expression of HOXA7-10 and the genes coding for MEIS1 and PBX3 in both NUP98::NSD1- and NUP98::TOP1-positive primary AML cells. NUP98::TOP1-cells showed early downregulation of HOXA7, 9, and 10 genes in the first week (Suppl. Figure S3A), whereas NUP98::NSD1-cells showed a significant reduction of HOXA gene expression in the second week of inhibitor treatment (Figure 3A), which is consistent with the kinetics of revumenib-mediated inhibition of cell proliferation. In contrast, expression of CDK6, a direct target gene of NUP98 fusion proteins, was markedly downregulated already at day 7 of treatment (Figure 3B). Primers are listed in the Supplemental Digital Content (Suppl. Table S4). We observed a dose-dependent downregulation of MEIS1 in all NUP98-r primary samples including a relapse sample (Figure 3C and 3D), while PBX3, IGF2BP2, and MEF2C expression were variably affected by revumenib (Figure 3E). Interestingly, expression of FLT3, a downstream target gene of MLL and MEIS1, was suppressed in a dose-dependent fashion in all NUP98-r samples with a more than 3-fold reduction at 250 nM of revumenib (Figure 3F). These data show that Menin inhibition can target FLT3 expression in line with similar observation reported for MLL-r and NPM1-mut AMLs.16.,27. Because all tested NUP98::NSD1 AML cells were positive for FLT3-ITD with allelic frequencies above 0.4, we also evaluated the impact of Menin inhibition on the expression of the mutated allele. We observed a 3-fold reduction of FLT3-ITD transcript levels upon exposure to revumenib (Figure 3G), suggesting that therapeutic modulation of the MLL–Menin interaction affects both NUP98 fusion- and FLT3-ITD-driven leukemic programmes. Nonetheless, the residual FLT3-ITD gene expression after treatment with revumenib may persist and be advantageous to leukemic cells. As there are reports indicating that incomplete eradication of this aberration in patients with FLT3-ITD+ leukemia may lead to relapse,30.,31. the moderate FLT3-ITD inhibition by revumenib raises the question if concomitant inhibition of the Menin–MLL interaction and FLT3-ITD signaling resulted in an even stronger reduction of leukemic propagation. In support of this hypothesis, it has been previously reported that the combined Menin and FLT3 inhibition exerts synergistic effect in MLL-r and NPM1-mut leukemias.27.,32. We therefore examined if the FLT3 inhibitor gilteritinib also synergises with revumenib in interfering with the expansion of NUP98::NSD1 FLT3-ITD+ patient-derived AML cells. FLT3 inhibition was found to be highly efficient against these AML cells (IC50= 30 nM), while primary AML cells (RUNX1::ETO) without FLT3 mutation were unaffected (Figure 3H). Considering that the Menin inhibitor (revumenib) has shown to suppress NUP98::NSD1 AML cells after 7–10 days, while FLT3-i induces rapid cytotoxicity, we treated AML cells with Menin inhibitor for 10 days followed by addition of FLT3-i for another 3 days. The combination of Menin and FLT3 inhibitors was highly synergistic with a combination index of 0.27 and strongly suppressed cell proliferation with significantly lower IC50 values of both drugs compared with single-drug treatment (Figure 3I). Taken together, although Menin inhibition alone impairs leukemic progression of NUP98-r/FLT3-ITD+ AML, the combination of Menin and FLT3 inhibitors increases their antileukemic efficacy in a synergistic manner.
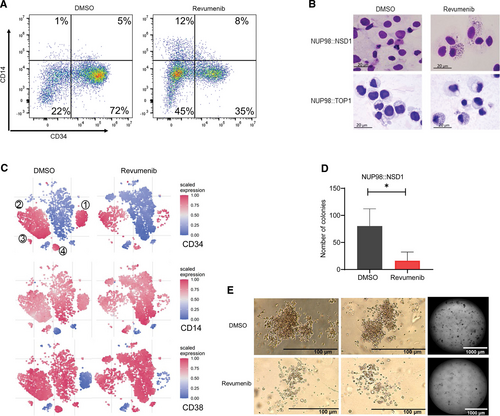
Menin inhibition induces differentiation of NUP98-r primary AML cells. (A) Representative flow cytometry data showing downregulation of CD34 and upregulation of CD14 expression in primary patient AML cells (NUP98::NSD1, FLT3-ITD+) upon treatment with revumenib (2500 nM). (B) Wright-Giemsa staining of primary AML cells 2 weeks after exposure to revumenib (250 nM). (C) t-SNE analysis showing distribution of subpopulations in 5 NUP98::NSD1 primary AML samples based on the expression of 8 cell surface markers. The t-SNE plots depict that immature subpopulations with CD34+ and CD38− markers decrease upon treatment with revumenib. The color is relative to the range of intensities in all cells. (D and E) Clonogenic potential of NUP98::NSD1 primary AML cells after treatment with revumenib (250 nM) as measured by a methylcellulose-based colony forming assay. Data were presented as mean (±SD). AML = acute myeloid leukemia; t-SNE = t-stochastic neighbor embedding.
Revumenib induces global gene expression changes in primary patient-derived NUP98::NSD1 AML cells
To characterize global gene expression changes in response to the inhibition of the MLL–Menin interaction, we performed RNA-sequencing with sorted AML cells from 3 patients after 7 and 12 days of exposure to revumenib or vehicle. Treatment with revumenib led to profound changes in gene expression pattern with 150 and 183 significantly (≥2-fold change, padj < 0.05) downregulated and upregulated genes, respectively (Suppl. Table S2). Extending the treatment time to 12 days increased the number of upregulated genes to 422 (Figure 4A and 4B; Suppl. Table S3). Genes downregulated at both time points include genes encoding key hematopoietic transcription factors (eg, MEIS1, PBX3, and MEF2C), receptor tyrosine kinases (FLT3 and KIT) and factors associated with stemness such as CD34, DNMT3B, and PROM1. Upregulated genes include members of the AP-1 transcription factor family (FOS, FOSB, and JUNB), inflammatory molecules (SGK1, CXCL2, and IL1R1) and factors involved in myeloid differentiation (eg, FGR, MMP8, and S100A8).
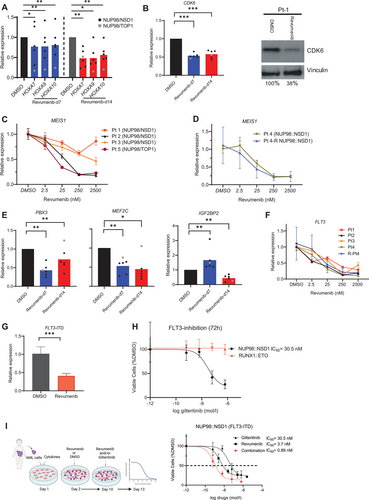
Menin inhibition suppresses the expression of NUP98 fusion target genes. Data were presented as mean (±SD). (A) qPCR results showing significant downregulation of NUP98 fusion target genes including HOXA7-10, (B) CDK6, (E) MEF2C, IGF2BP2, and PBX3 in NUP98-r primary AML samples upon treatment with revumenib (250 nM). (B) Suppression of CDK6 after 14 days of treatment was confirmed at protein level by western blot. (C, D, and F) qPCR results showing expression of MEIS1 and FLT3 after 7 days of treatment. (G) FLT3-ITD expression was downregulated after Menin inhibition as measured by qPCR with ITD-specific primers 1 week after treatment. (H and I) Dose-response curves of patient-derived NUP98::NSD1 (FLT3-ITD+) and RUNX1::ETO AML cells treated with gilteitinib, revumenib, or their combination. Dashed line represents IC50 value. AML = acute myeloid leukemia; qPCR = quantitative PCR.
Gene set enrichment analysis revealed a strong inverse correlation between the gene expression signature induced by Menin inhibition and the NUP98::HOXA9 fusion protein signature, further supporting the notion that interference with the MLL–Menin interaction directly affects the core transcriptional programme of NUP98 fusion proteins. Furthermore, there was a highly significant overlap of genes downregulated by menin inhibition between our study and Heikamp and colleagues20. (Suppl. Figure S4A and Suppl. Table S5). Moreover, the MLL–Menin inhibition was associated with loss of MYC-target genes, impaired cell cycle progression, loss of leukemic stemness, and higher H3K27me3 levels indicating increased EZH/PRC2 activity (Figure 4C and 4D). Overall, these data demonstrate that inhibition of the Menin–MLL interaction blocks leukemic self-renewal potential and reactivates myeloid differentiation in NUP98::NSD1 primary AML cells.
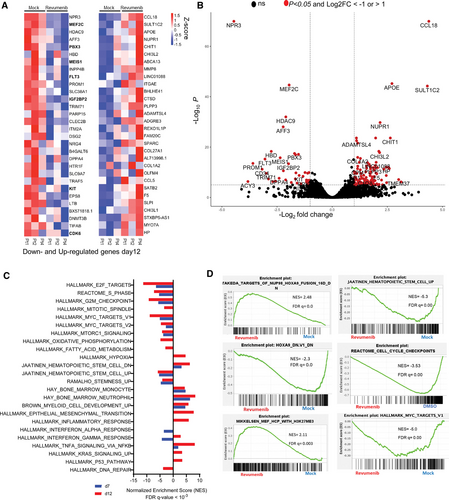
Gene expression changes of primary AML samples (NUP98::NSD1, FLT3-ITD+) associated with revumenib treatment. (A) Heat map of RNA-Seq analysis from 3 different primary patient samples (NUP98::NSD1, FLT3-ITD), representing top 30 differentially expressed genes 12 days after treatment with revumenib (250 nM). (B) Volcano plot illustrating differentially regulated genes in NUP98::NSD1 patients (n = 3) treated with revumenib (250 nM) compared with DMSO-treated samples (n = 3) on day 12 of treatment. Each dot represents a transcript with detectable expression. The FC (in log 2 scale) of the transcripts is plotted on the x-axis and the statistical significance expressed as −log10(FDR) on the y-axis. Genes not classified as differentially expressed are shown in black. (C and D) GSEA analysis of RNA-seq data showing enriched gene sets and pathways in revumenib (250 nM)-treated samples compared with DMSO-treated groups. AML = acute myeloid leukemia.
Revumenib eradicates leukemic cells in a PDX model of AML with NUP98::NSD1 (FLT3-ITD)
Next, we examined the dependency of NUP98-r AML engraftment and in vivo propagation on a functional MLL–Menin interaction. We therefore tested the efficacy of revumenib in a NUP98::NSD1 PDX mouse model (Figure 5A). NSG mice were intrafemorally transplanted with NUP98::NSD1+ FLT3-ITD+ PDX cells derived from patient 1. Revumenib treatment was initiated after confirming engraftment by continuous provision of chow containing 0.033% or 0.1% revumenib. Control animals showed progressing disease as indicated by increasing blast counts, whereas revumenib-treated animals experienced no increase in the number of peripheral blood leukemic cells over the whole observation period (Figure 5C). While the median survival of control animals was 147 days, all animals of the low dose revumenib group reached the end point of the study without showing clinical signs of disease. In the high dose group, 2 animals died without detectable leukemia, and 2 with pale bone marrow and splenomegaly (Figure 5B–5E). Postmortem inspection at the end of the study identified 1 additional animal from the low dose group with leukemic infiltration of the bone marrow and the spleen. Importantly, revumenib treatment completely eradicated leukemic blasts (hCD45+, hCD34+, and hCD33+) in the remaining 8 mice (Figures 5D and 5E). Taken together, these data show that revumenib has potent antileukemic activity toward NUP98::NSD1 AML even with FLT3-ITD as cooccurring mutation.
DISCUSSION
Here, we show that the phenotype and leukemic signature of NUP98-r AML cells are dependent on a functional MLL–Menin interaction in primary AML samples. Blocking this interaction with a Menin inhibitor interfered with leukemic transcriptional programmes associated with impaired proliferation, clonogenicity and facilitated myeloid differentiation, and prevented engraftment of NUP98::NSD1 FLT3-ITD patient cells in immunodeficient mice.
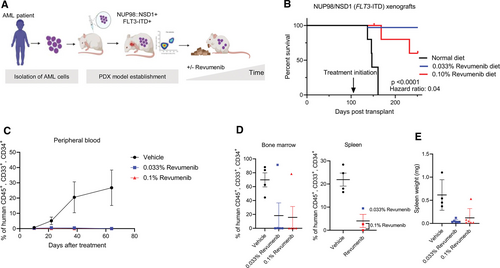
In vivo efficacy of revumenib in a NUP98::NSD1 (FLT3-ITD) AML PDX model. (A) Schematic representation of the in vivo study. (B) Kaplan–Meier survival plots showing the median survival upon treatment with revumenib or vehicle. Arrow indicates begin of treatment. (C) Flow cytometric quantification of human CD45+ cells in peripheral blood of NUP98::NSD1 FLT3-ITD PDX mice during treatment with revumenib or vehicle. Mean ± SE, n = 5 per group. (D) Flow cytometry data showing the frequency of leukemic blasts (hCD45+, hCD34+, hCD33+) in the spleen and bone marrow of NUP98::NSD1 (FLT3-ITD) PDX mice. (E) Spleen weight of NUP98::NSD1 (FLT3-ITD) PDX mice. AML = acute myeloid leukemia.
Previous studies addressing the effect of Menin inhibition in AML have used genetically engineered cell lines or characterized effects of Menin inhibition on NPM1-mutated, MLL-r or with NUP98-r mouse PDX models, demonstrating that the MLL–Menin interaction is a dependency in these AML types.16.,20.,26.,27.,33. We have now extended these investigations by examining the impact of MLL–Menin inhibition in primary patient AML cells expression NUP98 fusion proteins.20.,33.-35. Transgenic cell lines have been instrumental in investigating core mechanisms by which NUP98 fusion proteins drive leukemia.14.,20.,24.,36.,37. However, they do not reflect the evolutionary processes of leukemogenesis and the clonal heterogeneities linked to this process. Furthermore, the interplay of the major driver oncogene with secondary mutations that are present in patients and the interactions of AML cells with its niche cannot be adequately studied using these models, although these aspects are highly relevant for assessing drug response. Our ex vivo results extend previous findings on the effect of inhibiting MLL–Menin interaction in NUP98-r AML. The ex vivo evaluation of epigenetic inhibitors such as Menin inhibitors require culture conditions that preserve the differentiation status of different AML subpopulations for prolonged periods. Therefore, we examined the ex vivo effects of revumenib by using a coculture system that maintains and expands patient-derived primary AML cells for >2 weeks. Using this platform, we could demonstrate that revumenib induced myeloid differentiation and suppressed proliferation in 5 different primary NUP98::NSD1 AML samples but does not trigger apoptosis. Moreover, we also showed that this treatment resulted in marked changes in NUP98::NSD1-driven transcriptional networks that also included the downregulation of FLT3-ITD, which is a very common mutation in NUP98-r AML. FLT3 has previously been shown to be a direct transcriptional target of MEIS1/HOXA9, suggesting that inhibition of the MLL–Menin interaction targets FLT3 and possibly FLT3-ITD expression via downregulation of MEIS1/HOXA.27.,38.-40.
In AMLs where FLT3-activating mutations, and in particular FLT3-ITD, are present, FLT3 is a critical therapeutic target.41.,42. There are currently several FLT3 inhibitors in clinical use but achieving to long-term remission when it is used as a single agent is challenging.27.,42.,43. FLT3-ITD is a secondary genetic event in in our patient-derived NUP98-r AML and its signaling may also cooperate via STAT5 with MLL–Menin complexes in regulating gene expression.27. Here we showed that Menin inhibition suppresses FLT3-ITD expression and also increases this AML subtype susceptibility to FLT3 inhibitors. Additionally, we demonstrate that Menin inhibition single treatment is sufficient for suppressing NUP98-r AML in the presence of concomitant mutations such as FLT3-ITD and WT1; however, the combination of Menin and FLT3 inhibitors significantly reduce the antileukemic efficacy of each agent. Furthermore, these findings highlight the therapeutic interference with the MLL–Menin interaction as a paradigm for affecting both the primary and a secondary genetic event at the functional (NUP98 fusion protein) and at the transcriptional (FLT3-ITD) level. In a clinically relevant NUP98::NSD1 PDX model, we found that MLL–Menin inhibition can completely eradicate this aggressive AML subtype even when a concurrent FLT3-ITD mutation was present.
Using transduced cells, Thanasopoulou et al previously reported that the expansion of NUP98::NSD1/FLT3-ITD+ AML cells is accelerated by FLT3-derived signals. Our findings demonstrate that expression levels of FLT3-ITD and also cell proliferation decrease upon ex vivo revumenib treatment of patient-derived NUP98::NSD1/FLT3-ITD+ AML cells suggesting that the NUP98 fusion is the main driver of leukemia development.22.
In this study, we show that the NUP98-r AMLs with different fusion partners such as NSD1, TOP1, and DDX10 are sensitive to Menin inhibition. However, considering the diversity of fusion partners, which fall into distinct leukemia phenotypes such as AMKL (NUP98::JARID1A) and the variety of epigenetic regulatory domains in this AML type, more samples are required to be tested to determine if all NUP98-r AMLs are dependent on the MLL–Menin interaction. For example, AMLs expressing NUP98::KMT2A have been reported to not react to Menin inhibition.44. This AML subtype, unlike other NUP98-r AMLs, does not show elevated expression of HOX genes and therefore might follow alternative mechanism of leukemogenesis. This could be attributed to the absence of exon 1 in KMT2A within NUP98::KMT2A fusion, which encodes the Menin interaction domain. Further research is needed to determine whether wild-type KMT2A in this AML subtype has any defining role in sensitivity to Menin inhibition.
Heikamp et al recently demonstrated the responsiveness of the NUP98::NSD1 PDX mouse model, which was also WT1 mutant, to Menin inhibition. However, the study identified another PDX model with additional mutations (ASXL1, IDH1, BCORL1, WT1, and FLT3-ITD) that did not respond to Menin inhibition, indicating that certain mutations or their combinations may confer resistance to Menin inhibition.20. FLT3-ITD and WT1 are the most prevalent concurrent secondary genetic alterations in this AML subtype, and their cooccurrence with NUP98 fusions confers a poor prognosis.2.,3.,21.,22.,45.,46.
Our ex vivo and in vivo results with NUP98-r AML models suggest that samples expressing mutated WT1 or FLT3-ITD respond favorably to MLL–Menin inhibition, indicating that these mutations do not necessarily lead to treatment resistance. However, with respect to the BCORL1 mutation, recent studies have suggested that it dynamically competes with NUP98 fusions for transcriptional regulation of developmentally regulated, stem cell-associated genes as a component of the polycomb repressive complex (PRC) 1.1.47. Nonetheless, BCORL1 mutations are uncommon in NUP98-r AML and are poorly understood.
Overexpression of MEIS1/PBX3 is sufficient for transformation of mouse hematopoietic stem cells and driving AML in vivo, as previously reported.48. Moreover, elevated expression of HOXA9 and MEIS1 genes is associated with a number of hematological malignancies, suggesting that these genes are involved in the development of a variety of leukemias.49.-51. Patients with high MEIS1 expression have shorter survival times than those with low MEIS1, indicating that MEIS1 expression could be a prognostic factor even in AMLs with normal karyotype.50.,52. In this context, it is interesting to note that Menin inhibition has profound antileukemic effects in MLL-r, NPM1-mutant, and NUP98-r acute leukemias where MEIS1/HOXA9 is highly expressed. In contrast, AML cells expressing the RUNX1::ETO fusion protein do not execute a HOXA9/MEIS1 transcriptional programme and are not affected by Menin inhibitors, strongly suggesting a correlation between HOXA/MEIS1 expression and MLL–Menin dependence. While HOXA9 expression is moderately suppressed by Menin inhibition, MEIS1 is one of the first genes to be downregulated upon inhibition of the MLL–Menin interaction. Consequently, high MEIS1 overexpression may indicate susceptibility of a given AML toward Menin inhibition. Alternatively, comparative analyses of global gene expression patterns and coclustering with MLL-r, NPM1-mut, or NUP98-r AMLs may identify patients potentially benefitting from Menin inhibitors. Future evaluation of a wider range of AML samples will be required to examine these hypotheses.
Taken together, these data show that NUP98-r AMLs are highly susceptible toward Menin inhibition even in the presence of cooccurring mutations including FLT3-ITD and WT1. Consequently, they strongly support the inclusion of patients suffering of this AML subtype in the clinical evaluation of Menin inhibitors.
AUTHOR CONTRIBUTIONS
MR wrote and edited the article, performed the experiments, interpreted, and analyzed data. HB and ST performed the experiments and analyzed data. KS discussed the study concept and edited the article. RC performed the experiments. MA analyzed data. AKH performed the experiments. FG conceptualized the study, discussed the study concepts, and edited the article. GMM interpreted data. CMZ conceptualized the study, discussed the study concepts, and edited the article. OH designed, conceptualized, and supervised the study, discussed the study concepts, interpreted data, and edited the article.
DATA AVAILABILITY
Data sharing requests should be sent to Olaf Heidenreich.
DISCLOSURES
OH received research funding from Syndax. GMM is employee and shareholder of Syndax Pharmaceuticals. All the other authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
This work was supported by a KiKa programme grant (329) to OH and grants from the Austrian Science Fund (FWF, grants no. TAI-490 and P35628) to FG. ST is the recipient of a DOC fellowship of the Austrian Academy of Sciences at the University of Veterinary Medicine.