Germline RUNX1 Intragenic Deletion: Implications for Accurate Diagnosis of FPD/AML
Funding/support: This work was supported in part by French grant from the Ligue Nationale Contre le Cancer (équipe labellisée 2016 to HR).
Disclosure: The authors have indicated they have no potential conflicts of interest to disclose.
Authorship contributions: J-EM, SDB, J-BM, AB, SK-H, OC, and NA provided samples and clinical data, ND, RB, AM-R, VS, CM, RF, PB performed laboratory experiments and analyzed biological data; J-BM, HR, CP, ND, IA-D provided critical interpretation for this case; J-BM, ND, IA-D wrote the manuscript. All authors discussed and approved the manuscript.
In 2016, the WHO classification added a new category entitled “myeloid neoplasms with germ line predisposition” to acute myeloid leukemia (AML) classification, reflecting the increasing importance of these malignancies. Diagnosis of such patients is of clinical relevance, as they can benefit of specific clinical care including hematopoietic stem cell transplantation in first complete remission and genetic counseling. Genetic analysis is imperative to avoid the use of hematopoietic stem cell transplantation from an asymptomatic mutation carrier family donor. Thus, genetic testing for genes implicated in hereditary myeloid malignancies has to be done, whatever the age of the patient, in case of suggestive personal and/or family history, this is facilitated by the expanded use of next-generation sequencing. Among these hereditary malignancies, the familial platelet disorder with predisposition to acute myelogenous leukemia (FPD/AML) was first described in 1999.1 Since then, increasing number of families has been reported in the literature. Patients harbor mild to moderate thrombocytopenia and a risk of around 35% to develop AML or myelodysplasia. This disease (OMIM 601399) is due to germline genetic anomalies affecting the transcription factor RUNX1. Missense, nonsense, and frameshift mutations of RUNX1 have been reported as well as small and large deletions. At the time of AML diagnosis, the most frequent acquired genetic alteration implicates a second alteration of RUNX1 (mutation on the second allele, trisomy 21 with duplication of the mutated/deleted allele or loss of heterozygosity),2, 3 attesting to the importance of RUNX1 dosage in AML development.4
A 62-year-old male (Fig. 1, I-1) was hospitalized at Gustave Roussy Cancer Center for diagnosis of an AML in August 2009. He had a history of mild thrombocytopenia unexplored for decades. He did not present any other clinical phenotype. His 2 children (II-1 and II-2) and one of his granddaughter (III-2) presented also a thrombocytopenia (II-1: 100 G/L, II-2: 80 G/L, III-2: 115 G/L) with history of easy bruising. The patient was diagnosed with an AML-M0 with normal cytogenetic, wild type for NPM1 and FLT3-ITD mutations. He was in complete remission after a 3 + 7 induction therapy (daunorubicin 60 mg/m2, Day 1 to Day 3 + cytarabin 200 mg/m2, Day 1 to Day 7) complicated with an invasive pulmonary aspergillosis, followed by 4 ambulatory postremission chemotherapy (45 mg/m2 daunorubicin, Day 1 + 60 mg/m2/12 h cytarabine as home subcutaneous infusions, Day 1 to Day 5) and a cord blood transplant. Unfortunately he relapsed and died in April 2011. The family and clinical history of the patient suggested inherited myeloid neoplasms associated with platelet disorders.
The biological samples were collected after informed consent, in accordance with the Declaration of Helsinki. DNA was extracted from cell pellets using the QIAamp Tissue Kit (Qiagen) according to the manufacturer's instructions.
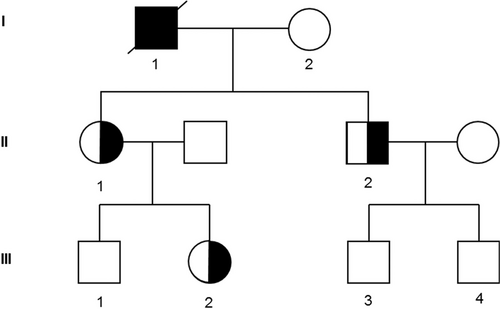
Family tree of the affected pedigree. The open symbols indicate pedigree members without thrombocytopenia or AML; the half black symbols, members with thrombocytopenia alone; the full black symbols, members with thrombocytopenia and AML. AML = acute myeloid leukemia.
Direct sequencing of all coding exons of the RUNX1 gene was performed as previously described by Sanger sequencing.3
For high-throughput sequencing (HTS), a panel of 36 genes recurrently mutated in myeloid malignancies was sequenced on genomic DNA isolated from mononuclear white blood cells. These include ASXL1, BCOR, BCORL1, CALR, CBL, CSF3R, DNMT3A, ETV6, EZH2, FLT3, GATA2, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NIPBL, NPM1, NRAS, PHF6, PTPN11, RAD21, RIT1, RUNX1, SETBP1, SF3B1, SMC1A, SMC3, SRSF2, STAG2, TET2, TP53, U2AF1, WT1, ZRSR2 genes. The library was prepared using the Ampliseq System according to the manufacturer's instructions and run on Ion Proton (Thermo Fisher, Waltham, MA). Raw data were analyzed with both Torrent Browser (Thermo Fisher) and SeqNext (JSI Medical System, Los Angeles, CA). A high depth of coverage (>1500) was obtained for all genes. A second panel of genes implicated in inherited myeloid malignancies including ANKRD26, CEBPA, DDX41, ETV6, GATA2, RUNX1, SRP72, TERC, TERT, and TP53 was sequenced according to the same protocol.
To detect RUNX1 deletions, SNP-array karyotyping (CytoScan HD array; Thermo Fisher Scientific) was performed according to manufacturers’ instructions. Data were analyzed with the Chromosome Analysis Suite version 3.1 (ChAS, Thermo Fisher Scientific). The intragenic RUNX1 deletion was then confirmed by multiplex ligation probe amplification (MLPA) (SALSA MLPA P437-A1 Familial MDS-AML probemix, MRC Holland).
Based on patient's personal and familial history of thrombocytopenia, we first sequenced RUNX1 by the Sanger method at AML diagnosis and discovered a RUNX1 mutation (NM_001001890 exon 2 c.416G>A:p.R139Q). The R139Q mutation has been already described in FPD/AML patients.1, 5 It affects the Runt homology domain (RHD) of RUNX1, which binds to DNA and to the other member of the CBF complex, CBFβ. After genetic counseling, we sequenced RUNX1 in the 2 adult's relative (II-1 and II-2) presenting thrombocytopenia but unexpectedly, we were not able to find the R139Q mutation or any other RUNX1 mutations. A clinical and biological follow-up was decided for the relatives. In 2016, we used a 36 genes-HTS panel to study the AML diagnosis sample of the proband. It confirmed the RUNX1 R139Q mutation with a variant allele frequency (VAF) at 50%. Additional somatic mutations were described as BCOR p.K1137delinsTX (VAF 54%), DNMT3A p.W795C (VAF 48%), NRAS p.Q61K (VAF 47%), and SRSF2 p.P95R (VAF 47%). Another HTS panel, including among others ANKRD26 and ETV6 which are also implicated in inherited myeloid malignancies with preexisting platelet disorders,6-9 was used but we did not find any additional mutation. Based on our recent paper describing a second aberration of RUNX1 in AML transformation in FPD/AML patients,2 we hypothesized that the RUNX1 mutation found at the AML stage was an acquired event and that we may have missed the inherited RUNX1 alteration.
As germline RUNX1 deletions have been already described in FPD/AML,1, 3, 10-14 we performed SNP-array and we were able to identify a microdeletion of 51 kb in the RUNX1 gene. This result was confirmed by MLPA in the patient and in the 2 adult's relative confirming the diagnosis of FPD/AML (Fig. 2A-B). The deleted region (aminoacids 143–251) encompasses parts of the exons 3 and 4 of RUNX1b isoform, which encodes parts of the RHD and transactivation domains (Fig. 2C). Interestingly, a frameshift mutation in the exon 4 of RUNX1, resulting in a premature termination codon 22 nucleotides downstream, has already been reported in FPD/AML (RUNX1 p.T219RfsX8) and is associated with AML development.2, 15 This suggests that such truncated mRNA could predispose to leukemia progression in FPD/AML. Besides large RUNX1 deletions, small deletions have been rarely described in FPD/AML, they were identified by MLPA or by CGH-array, confirming once again the importance of this type of genetic test for FPD/AML diagnosis.11, 12 Additionally, we sequenced the proband's sample at complete remission using the 36 genes-HTS panel and did not find the mutations identified at AML stage, excepted the DNMT3A mutation with a lower VAF at 4%, which could represent the persistence of clonal hematopoiesis. We were not able to find the RUNX1 mutation anymore, confirming the acquired status of this genetic event.
This report illustrates that, in front of a clinical and familial history evocating strongly inherited thrombocytopenia with predisposition to AML, all the biological examinations have to be conducted. Indeed, it is crucial to diagnose inherited diseases such as FPD/AML, as the management of the patient could differ from a patient with de novo AML, especially if hematopoietic stem cell transplant is considered. Moreover, it implicates a regular follow-up for patients within the family at thrombocytopenia stage, as they are at risk of developing AML. In our case, Sanger sequencing and amplicon-based HTS were not sufficient to establish the diagnosis but allowed to identify an acquired second hit affecting RUNX1 at AML stage, confirming the association of this event with AML development. Of note, capture-based HTS may provide information on copy number variations for all homogeneously covered regions. This notion should be kept in mind in lab’ strategies especially for the assessment of haplo-insufficient genes status. In FPD/AML, as both deletions and mutations of RUNX1 have been reported in patients, biological tests should include other techniques such as SNP-array and/or MLPA in a second phase to identify microdeletions when FPD/AML is strongly suspected. Similar strategy should be applied to diagnose other inherited hematological disorders implicating not only mutations but also gene deletions.
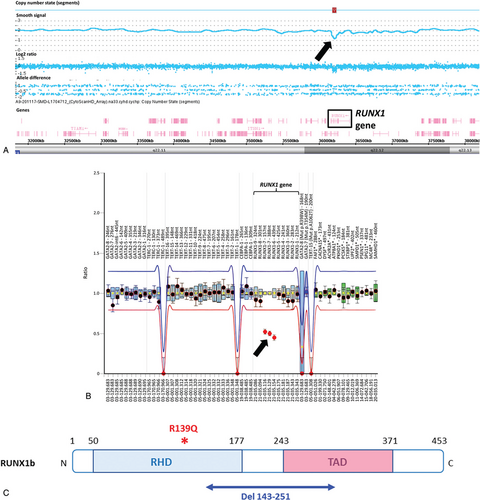
RUNX1 alterations identified in the family. (A) SNP-array karyotyping. View of the chromosome 21 (coordinates chr21: 31,803,844–38,217,464). The black arrow shows the deletion within the RUNX1 gene shared by all 3 individuals (I-1, II-1, and II-2). Picture is from Chromosome Analysis suite. (B) Ratio chart of MLPA analysis. A ratio of 1 indicates no differences in respect to reference samples, while a ratio of 0.5 indicates a heterozygous deletion. Dots represent MLPA probes and lines indicate thresholds. The 3 dots, located below the lower threshold (black arrow), indicate the RUNX1 intragenic deletion shared by all 3 individuals (I-1, II-1, and II-2). Notice that numbering of the exons 6 and 7 in the MLPA analysis corresponds to exons 3 and 4 of the RUNX1b isoform (NM_001001890). (C) Localization of RUNX1 abnormalities. The isoform b is represented. Germline alteration of RUNX1 corresponding to the deletion of the aminoacids 143 to 251 (in blue) affects part of the RHD and TAD domains, whereas the acquired RUNX1 mutation p.R139Q (in red) is located on the RHD domain, upstream of the deleted region. MLPA = multiplex ligation probe amplification, RHD = Runt homology domain, SNP = single nucleotide polymorphism, TAD = transactivation domain.
Acknowledgments
The authors thank the patients for their participation. This work was supported in part by French grant from the Ligue Nationale Contre le Cancer (équipe labellisée 2016 to HR). The authors thank the Tumor Bank of Lille Hospital (certification NF 96900-2014/65453-1) for handling, conditioning, and storing the patient's samples.