P596: DELETION OF THE SHORT ARM OF CHROMOSOME 8 AND TNFRSF10B LOSS ASSOCIATE TO POOR PROGNOSIS AND DRUG RESISTANCE IN CHRONIC LYMPHOCYTIC LEUKEMIA
Background: Chronic lymphocytic leukemia (CLL) is a very heterogeneous disease whose prognosis varies according to cytogenetics.
Aims: We performed a precise characterization of a rare abnormality associated with aggressive CLL, the deletion of the short arm of chromosome 8 (del8p).
Methods: We correlated patients’ cytogenetic and clinical outcomes using standard statistical methods. We performed in vitro analyses on primary CLL cells and on TNFRSF10B CRISPR/Cas9 edited OSU-CLL cell lines using flow cytometry for drug response assays and ddPCR for genes expression.
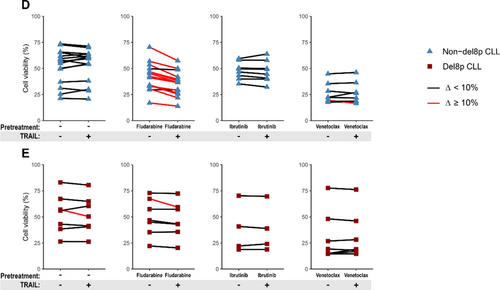
Results: Comparing the largest series to date of del8p CLL (n=57) to a non-del8p cohort (n=155), del8p was significantly associated with poor prognostic factors: complex and highly complex karyotypes (HCK) (72% and 44%, vs 22% and 8% respectively, p<.001), del11q (52% vs 17%, p<.001), del17p (36% vs 10%, p<.001) and unmutated IGHV (73% vs 46%, p=.002). Del8p CLL had a shorter time to first treatment (TTFT, median 0.25y vs 1.7y, p=.007), a shorter overall survival (OS 10y: 30% vs 82%; median 6.8y vs not reached, p<.0001), and a higher risk of Richter transformation (RT cumulative incidence 10y: 22% vs 5.5%, p=.034). Additionally, large del8p and TP53 disruption negatively affected the OS. Interestingly, regarding CLL treated with fludarabine-based regimens, del8p CLL had a shorter next treatment-free survival (10y: 3.7% vs 21.5% without new treatment, p=.002) and a shorter OS (10y: 45.9% vs. 81.6%, p<.001) compared to non-del8p CLL, including CLL with mutated IGHV. One copy of the TNFRSF10B gene, coding a pro-apoptotic receptor activated by TRAIL, was lost in 91% of del8p cases. This is key because TNFRSF10B was haploinsufficient in del8p cells and was involved in the modulation of fludarabine-induced cell death, as confirmed in primary CLL and cell lines. (1) The expression of TNFRSF10B was significantly lower in del8p than in non-del8p cells (median ratio 1.52 vs 3.66; 2.41-fold decrease, p<.001). (2) TNFRSF10B was functional in inducing cell death (PCD) in cell lines. (3) TRAIL alone being not sufficient to induce PCD in primary CLL cells, we hypothesized that exposure of cells to drugs might modulate TNFRSF10B expression, resulting in improved sensitivity to TRAIL: (i) in non-del8p cells, TNFRSF10B expression was significantly increased after exposure to fludarabine (2.8-fold), but not after ibrutinib or venetoclax. This increase was not observed in del8p cells; (ii) we performed a sequential approach to evaluate the cytotoxic effect of TRAIL with fludarabine. In non-del8p cells, fludarabine + TRAIL resulted in a significant loss of cell viability, and TRAIL had an additional cytotoxic effect in 13/15 (87%) samples (Fig 1D). By contrast, this sequential regimen failed to elicit a synergistic cytotoxic effect in 6/7 del8p samples, revealing that del8p avoids the synergy between fludarabine and TRAIL-induced PCD (Fig 1E).
Summary/Conclusion: Del8p is associated with poor prognostic factors (CK, HCK, TP53 aberrations, unmutated IGHV), short TTFT and OS, and RT. Additionally, we demonstrate that TNFRSF10B plays an important role in fludarabine resistance in CLL, in vivo and in vitro. Our results argue for the assessment of del8p before choosing a fludarabine-based therapy.
Keywords: Chronic lymphocytic leukemia, TRAIL, Chromosomal abnormality, Gene deletion