Individual and Common Antigen-Recognition Sites of Liver-Derived T Cells in Patients with Autoimmune Hepatitis
Abstract
Autoimmune hepatitis (AIH) is characterized by dense T-cell infiltrations in the liver tissue, but little is known how T cells influence the pathogenesis. To address this question, the distribution of T-cell receptor variable β-chain (TCR Vβ) transcripts of peripheral blood and liver-infiltrating T cells from previously untreated patients with newly diagnosed acute exacerbated AIH was investigated. Furthermore, the lengths and sequences of complementary-determining region 3 (CDR3) were studied. Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis and CDR3 spectratyping revealed multiple clonal expansions of liver-infiltrating T cells but not peripheral T cells within various TCR Vβ families. Further analysis of overexpressed TCR Vβ transcripts using TCR β-chain-joining element (TCR Jβ)-specific primers in a nested PCR showed characteristic Vβ/Jβ combinations. Subsequent sequencing of CDR3 regions from PCR products confirmed the clonality of T-cell expansions and the usage of common and individual CDR3 motifs. In conclusion, the clonality of expanded T cells within the liver tissue during early clinical manifestation of untreated AIH indicated that autoantigen-specific T cells accumulate at the inflammation site. Individual and common CDR3 motifs argued for predominant epitopes that were recognized by liver-infiltrating T cells in AIH patients.
Introduction
Autoimmune hepatitis (AIH) is a liver-specific autoimmune disease characterized by dense T-cell infiltrations in the liver, serum autoantibodies and genetic predisposition of HLA-DR3 in European patients or HLA-DR4 in Japanese patients [1–3]. Recently, the asialoglycoprotein receptor and a recombinant microsomal antigen from liver and kidney (LKM-1) have been described as antigens of liver-infiltrating T cells in AIH patients and represent candidate autoantigens that may trigger the disease process [4–6]. Infiltrating T cells that predominantly belong to the CD4+ helper cells and express the T-cell receptor (TCR) α- and β-chains are suggested to contribute to the immunopathogenesis of AIH, but their role herein is unclear [7, 8].
Via their specific TCR, these T cells could recognize autoantigens presented by major histocompatibility complex (MHC) class I or class II molecules and then undergo clonal expansion [9]. The accumulation of T cells with a limited repertoire of variable regions within the TCR β-chains (TCRBV) was demonstrated in several autoimmune diseases, like experimental autoimmune encephalomyelitis, multiple sclerosis and reactive arthritis [10–12]. Using reverse transcriptase-polymerase chain reaction (RT-PCR) and subsequent single-strand conformation polymorphism (SSCP) analysis, the accumulation of many oligoclonal T cells was demonstrated in the liver tissue of AIH patients [13]. Furthermore, the clonotypic analysis of liver-infiltrating T cells revealed an Asp–Arg–Pro motif in the complementary-determining region 3 (CDR3) of TCR variable β-chain 3 (TCR Vβ3) in three HLA-DR4-positive Japanese AIH patients. In European AIH patients, TCR Vβ3, Vβ5.2 and Vβ13.1 molecules were overexpressed in liver-infiltrating T cells [14]. Furthermore, it was demonstrated that LKM-1 peptide-specific T-cell clones from different patients with type II AIH shared a common motif within their CDR3 [15]. The high-sensitive analysis using spectratyping and nucleotide sequencing of CDR3 revealed individual T-cell expansions but no T-cell clones with identical CDR3 motifs between different HLA-DR4-positive Japanese patients [16, 17].
In this study, molecular analysis of liver-infiltrating and peripheral blood T cells should be performed to detect characteristic clonal T-cell expansions into the livers during the early manifestation of AIH in previously untreated European patients.
Materials and methods
Patients One male and two female patients with acute exacerbations of newly diagnosed AIH were enrolled in this study. All patients fulfilled the diagnostic criteria of the International Autoimmune Hepatitis Group and showed high inflammatory activity in diagnostic liver biopsies [18]. None of the patients had serological markers for chronic viral hepatitis B or C or human immunodeficiency virus. At the time of the study, none of the patients received an immunosuppressive therapy. HLA typing was performed by standard microtoxicity technique (HLA class I and class II molecules) and PCR-restriction fragment length polymorphism (RFLP) technique (HLA-DRB alleles) [19]. Clinical patient data are presented in Table 1.
| Patient | Sex | Age (years) | Autoantibodies | γ-Globulins (%)* | IgG (g/l)† | ALT (U/l) | GGT (U/l) | HLA |
|---|---|---|---|---|---|---|---|---|
| 1276 | F | 47 | ANA SMA | 579 | 234 | A2,3, B47,60, Cw3,6 DR1,4, DQ5,8, DR53 | ||
| 1376 | F | 33 | SMA | 25.4 | 954 | 210 | A1,3, B7,8, Cw7, DR3, DQ2, DR52 | |
| 1488 | M | 77 | ANA | 22.5 | 690 | 254 | A1,2, B8,45, Cw6,7, DR3,7, DQ2 |
- ALT, alanine aminotransferase; GGT, γ-glutamyltransferase; HLA, human leucocyte antigen; ANA, antinuclear antibody; SMA, smooth muscle antibody.
- * γ-Globulins in serum electrophoresis (% of total protein).
- † Quantitative analysis of immunoglobulin G (IgG, g/l).
All patients gave their written informed consent in accordance with the Helsinki Declaration of Ethical Guidelines.
Liver-infiltrating and peripheral blood T cells Small pieces of diagnostic liver biopsies (<2 mm) were incubated for 48 h in 96-well flat-bottom microtitre plates in culture medium (RPMI-1640) supplemented with human AB serum and 20 U/ml of interleukin-2, as described recently [6]. Liver-infiltrating T cells were collected and washed for 10 min at 200 g at room temperature. Peripheral blood mononuclear cells were collected from the interphase after Ficoll density centrifugation and washed as described above.
RNA isolation and cDNA preparation Viable cells were separated from dead cells and detritus by another Ficoll density centrifugation (50% Ficoll 1077 and 50% Ficoll 109; Pharmacia, Freiburg, Germany), followed by two washings. Then, cell pellets were homogenized with 350 µl of lysis buffer containing guanidine isothiocyanate and 0.1% mercaptoethanol. After the addition of 350 µl of 70% ethanol, 700 µl was loaded on spin columns and centrifuged for 15 s at 14,000 g. Nucleic acids were purified using RW1 wash buffer (700 µl), 2× R-phycoerythrin wash buffer (500 µl) and maximum-speed centrifugation for 2 min each. Then, spin columns were set on new 1.5 ml tubes, and RNA was eluted by 2× addition of 50 µl of diethyl pyrocarbonate and H2O for 1 min at 14,000 g, followed by the addition of 1 µl of RNase inhibitor.
The reverse transcription reaction was initiated with 4 µl of oligo (dT) primers and 18 µl of RNA extract for 5 min at 70 °C. Then, 8 µl of single-strand buffer (5×), 4 µl of 100 mm dithiothreitol, 2 µl of 2.5 mm dNTP, 2 µl of mouse moloney leukemia virus (MMLV) (200 U/l; Gibco BRL, Grand Island, NY, USA) and 1 µl of RNase inhibitor were added and heated for 1 h at 42 °C and for 5 min at 94 °C. For PCR reaction, 1 : 20 dilutions were used.
TCR V β-specific PCR amplification cDNA samples diluted 1 : 20 were used for PCR amplification using a TCR constant region primer (Cβ) and 26 TCR Vβ family-specific primers [14]. Glyceraldehyde phosphate dehydrogenase-specific primers and H2O served as internal controls. In brief, each reaction mixture contained 14.56 µl of H2O, 3.5 µl of 10× buffer, 3.5 µl of Cβ primer, 2.8 µl of dNTP and 0.14 µl of Taq polymerase (Perkin Elmer, Freiburg, Germany), together with 7 µl of cDNA and 3.5 µl of Vβ-specific primers in a single tube. Vβ-specific cDNA was amplified by 34 cycles in a Hybaid thermocycler (MWG Biotech, Langen, Germany), starting with denaturation at 94 °C for 1 min, followed by annealing at 60 °C for 1 min and extension at 72 °C for another 1.5 min. PCR amplification was stopped finally by 10 min at 72 °C. Then, 10 µl of each amplified PCR product was separated on a 2% agarose gel containing ethidium bromide.
TCR β-chain-joining element (TCR J β)-specific nested PCR amplification Nested PCR with 13 Jβ-specific primers was performed with the amplified Vβ-specific PCR products. In brief, the master mix contained 13.3 µl of individual Vβ PCR product, together with 40 µl of 10× buffer, 32 µl of dNTPs, 233 µl of H2O and 1.6 µl of Taq polymerase. Then, 37.5 µl of this mixture was added to 2.5 µl of individual Jβ-specific primers. H2O served as negative control and Cβ primer reaction as positive control. After 36 cycles of amplification, as described above, the PCR products were separated on a 2% agarose gel.
Spectratyping (CDR3 length assay) CDR3 lengths were analysed with PCR products from Cβ/Vβ PCR. cDNA copies of 0.1 µg of total RNA were amplified by Cβ/Vβ nested PCR in 50 µl total volume for 36 cycles. In selected cases, spectratyping was also performed with fluorochrome-labelled 13 Jβ primers using the Cβ/Vβ PCR products as templates [16, 17]. About 6 µl of aliquots of the PCR products plus 3 µl of stop solution were denatured for 5 min at 90 °C. Then, the PCR products were separated on 6% polyacrylamide denaturing sequencing gels containing 8 m urea for 2 h at 40 mA. For fixation, the gels were incubated overnight in 10% acetate before silver staining was performed to visualize the bands. Furthermore, CDR3 lengths were determined by spectratype fragment plot analysis in an automated sequencer. Each master mix contained 2 µl of PCR product, 2 µl of 10× buffer, 1.6 µl of dNTPs, 10.3 µl of H2O and 0.1 µl of Taq polymerase, together with 2 µl of Vβ primers (2 µm) and 2 µl of Cβ primers (2 µm). PCR products were copied by five cycles in a single tube, starting with denaturation at 94 °C for 1 min, followed by annealing at 60 °C for 1 min and extension at 72 °C for another 1.5 min. PCR amplification was stopped finally by 10 min at 72 °C, and the PCR products were separated on 6% polyacrylamide gels in an ABI 377 sequencer (Applied Biosystems, Inc, Foster City, CA, USA). After the gels had been stained, CDR3 sizes of the fragments were determined automatically by gene scan 672 software.
DNA sequencing After spectratyping, bands of interest were cut from the polyacrylamide sequencing gels and transferred to 1.5 ml caps. cDNA fragments were eluted using 200 µl of Maxam–Gilbert buffer containing 0.5 m ammonium acetate, 10 mm magnesium acetate, 1 mm ethylenediaminetetraacetic acid, pH 8.0, and 0.1% sodium dodecyl sulphate overnight and purified with 2 volumes of 100% ethanol and 1/10 volume of 3 m sodium acetate (pH 5.0) at 20,000 g for 45 min at 4 °C. Then, the pellets were washed with 70% ethanol at 14,000 r.p.m. for 15 min at 4 °C DNA, dried overnight and dissolved in 75 µl of H2O before the DNA fragments were analysed 1 : 10 by PCR.
After ligation of 2 µl of PCR product with 2 µl of pPCR 2.1, 1 µl of T4-DNA ligase, 1 µl of 10× buffer and 4 µl of H2O overnight at 13 °C, the PCR products were transformed into Escherichia coli (Invitrogen, Carlsbad, CA, USA). In brief, 2 µl of the ligated PCR product was mixed with 2 µl of β-mercaptoethanol and incubated for 30 min on ice, 30 s in a 42 °C water bath and cooled again for 2 min on ice. After 250 µl of SOC medium was added and vortexed for 1 h at 37 °C at 220 r.p.m., the transformed bacterial solution was incubated in X gel-Amp LB plates overnight at 37 °C. White colonies with inserts were picked and expanded using the Qiagen miniprep kit.
Inserts were cut using EcoRI restriction enzyme, and plasmids each representing a cloned PCR product underwent a sequencer reaction. Nucleotide sequences were determined by the dideoxy method using the ABI 377 automated sequencer.
Results
To detect overexpressed TCR Vβ molecules in untreated AIH patients, RT-PCR analysis was performed with cDNA isolated from liver-infiltrating T cells and corresponding peripheral blood T cells. Using a Cβ-specific primer and 26 TCR Vβ-specific primers and subsequent semiquantitative analysis by densitometry of the agarose gels, multiple TCR Vβ molecules were found overexpressed by liver-infiltrating but not by peripheral blood T cells in the three tested AIH patients. In detail, Patient 1276 showed overexpressed TCR Vβ2, 3, 4, 5.2, 6 and 21 molecules and significantly reduced expressions of TCR Vβ molecules 8, 23 and 24 in the liver-infiltrating T cells compared with that in autologous peripheral blood T cells. Patient 1367 strongly overexpressed the TCR Vβ6 molecule and had reduced expression of the TCR Vβ21 molecule in her liver. In Patient 1488, finally, the expression rates of the TCR Vβ2, 4 and 5.2 molecules were significantly enhanced, while the expression rates of the TCR Vβ8, 15, 21 and 23 molecules were reduced in the liver-infiltrating T cells compared with that in autologous peripheral blood T cells.
To demonstrate potential accumulation of single T-cell clonotypes, liver-infiltrating and peripheral T cells were further analysed by CDR3 size spectratyping. In the peripheral blood, most PCR transcripts showed Gaussian distributions of CDR3 lengths, indicating the polyclonality of T cells. However, liver T cells of all the three AIH patients showed multiple characteristic spectratypes with one or two overexpressed bands, each representing the usage of distinct CDR3 lengths by T-cell clonotypes. In detail, common patterns of CDR3 lengths were observed within Vβ3-, 4-, 5.2-, 9-, 11-, 13.1-, 14-, 16- and 22-specific PCR transcripts. In addition, individual T-cell expansions were found in Patient 1276 with Vβ2- and 17-specific PCR products, in Patient 1367 with Vβ8- and 17-specific PCR products and in Patient 1488 with Vβ1- and 18-specific PCR products (Fig. 1).
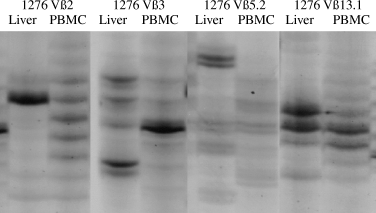
Spectratyping was performed to analyse the lengths of the complementary-determining region 3 (CDR3) regions of liver-infiltrating (Liver) and peripheral blood T lymphocytes (PBMC) from representative patient with autoimmune hepatitis (Patient 1276). Single and strong bands indicate the preferential usage of identical CDR3 lengths, a prerequisite of T-cell clonotypes. Vβ2, Vβ3, Vβ5.2 and Vβ13.1 overexpressed T-cell receptor molecules.
Furthermore, the plot fragment analysis revealed one or two peaks, indicating characteristic CDR3 length profiles of selected Vβ-specific PCR transcripts from liver-infiltrating T cells that were not observed with peripheral blood T cells (Fig. 2).
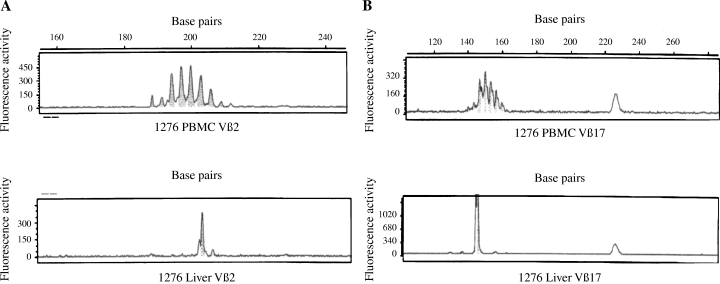
Fragment plot analysis of liver-infiltrating (Liver) or peripheral blood T cells (PBMC) expressing the T-cell receptor variable β-chain 2 (TCR Vβ2) (A) or Vβ17 (B) molecules in representative Patient 1276 with autoimmune hepatitis. The non-Gaussian distribution of polymerase chain reaction transcripts in the liver indicates the usage of distinct complementary-determining region 3 (CDR3) lengths and therefore the clonality of the T cells.
In the next set of experiments, PCR products from liver-infiltrating T cells with overexpressed Vβ molecules or peaks in their CDR3 size distribution were further analysed by nested PCR using 11 Jβ-specific primers. In Patient 1276, transcripts specific for the combinations Vβ14 with Jβ1.2 or 2.4, Vβ2 with Jβ2.4 or 2.5, Vβ11 with Jβ2.7 and Vβ22 with Jβ2.5 showed strong and single bands in the gel electrophoresis (Fig. 3).
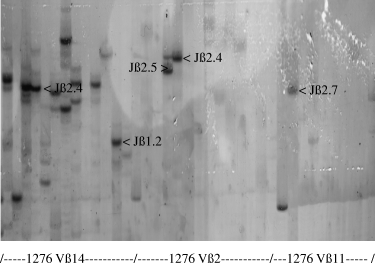
Polymerase chain reaction (PCR) transcripts specific for T-cell receptor variable β-chain (TCR Vβ) molecules derived from liver-infiltrating T cells of Patient 1276 were analysed by nested PCR using primers for 13 TCR Jβ and spectratyping. Each band represents the usage of a single Jβ element in combination with a single Vβ element.
The analysis of the other AIH patients revealed common usage of the Vβ/Jβ combinations: Vβ22 with Jβ2.5, Vβ11 with Jβ2.5 or 2.6 and Vβ14 with Jβ1.2, 1.3 or 2.4 molecules in Patient 1367, and Vβ11 with Jβ2.5, 2.6 or 2.7 and Vβ22 with Jβ2.5 molecules in Patient 1488.
After nested PCR and gel electrophoresis, selected bands were cut from the agarose gels. Then, PCR products specific for distinct Vβ and Jβ molecules were purified and cloned before they had been sequenced to characterize the N–D–N diversity regions within the antigen-recognition sites. Sequence analysis performed with multiple cloned PCR transcripts derived from liver-infiltrating T cells (Patient 1276, n = 25; Patient 1367, n = 10; and Patient 1488, n = 11) revealed that the vast majority of PCR products derived from an individual AIH patient with identical Vβ/Jβ combinations shared identical or very similar antigen-recognition sites with the replacement of single amino acids. Furthermore, sequence homologies within the CDR3 motifs of liver-infiltrating T cells from different AIH patients expressing the same Vβ/Jβ combinations were observed: for example, XXXXXXX(XX)GANVL in Vβ3/Jβ2.6-expressing clones from Patients 1276 and 1488, SE(XX)A(X)ETQ in Vβ11/Jβ2.5-expressing clones from Patients 1367 and 1488 and SP(X)G(X)QNSPL in Vβ18/Jβ1.6-expressing clones from Patients 1276 and 1367. Identical CDR3 motifs indicating the recognition of identical epitopes on the unknown autoantigens have not been observed. Table 2 summarizes the detailed data of the sequence analysis.
| Patient | Vβ/Jβ | Clone | Vβ | N–D–N | Jβ |
|---|---|---|---|---|---|
| 1276 | 3/2.6 | K1 | CAS | TPGDGSNEAGANVL | TFG |
| 3/2.6 | K2 | CAS | TPGDGSNEAGANVL | TFG | |
| 1488 | 3/2.6 | K5 | CAS | TTTGFNSGANVL | TFG |
| 3/2.6 | K6 | CAS | ITTGFNSGANVL | TFG | |
| 1376 | 11/2.5 | K1 | CAS | SEGPAGETQ | YFGPGTRLLV |
| 11/2.5 | K2 | CAS | SEGPAGETQ | YFGPGTRLLV | |
| 1488 | 11/2.5 | K5 | CAS | SEAETQ | YFGPGTRLLV |
| 11/2.5 | K6 | CAS | SEAETQ | YFGPGTRLLV | |
| 1276 | 13.1/2.5 | K1 | CAS | SSPKRGGTQ | YFGPGTRLLV |
| 13.1/2.5 | K2 | CAR | LAGDLKGETQ | YFGPGTRLLV | |
| 1488 | 13.1/2.5 | K1 | CAS | SGGEQETQ | YFGPGTRLLV |
| 13.1/2.5 | K2 | CAS | SSFLAIEDQ | YFGPGTRLLV | |
| 13.1/2.5 | K2 | CAS | SSFLAIETQ | YFGPGTRLLV | |
| 1276 | 14/1.2 | K1 | CAS | SLKGANYGY | TFGSGTR |
| 14/1.2 | K2 | CAS | SLNVDDYGY | TFGSGTR | |
| 1376 | 14/1.2 | K1 | CAS | SPMGAGEAGY | TFGSGTR |
| 14/1.2 | K2 | CAS | SPMGAGEAGY | TFGSGTR | |
| 1276 | 14/2.4 | K1 | CAS | RPTRGGNIQ | YFGAGTRLSVL |
| 14/2.4 | K2 | CAS | RPTRGGNIR | YFGAGTRLSVL | |
| 1376 | 14/2.4 | K1 | CAT | ARGGLENIQ | YFGAGTRLSVL |
| 14/2.4 | K2 | CAT | ARGGLENIQ | YFGAGTRLSVL | |
| 1276 | 18/1.6 | K2 | CAS | SPQGFQNSPL | HFGNGTRLTV |
| 18/1.6 | K4 | CAS | SPQGFQNSPL | HFGNGTRLTV | |
| 1376 | 18/1.6 | K1 | CAS | SSRGQDNSPL | HFGNGTRLTV |
| 18/1.6 | K4 | CAS | SPLVRGNSPL | HFGNGTRLTV | |
| 1276 | 22/1.1 | K1 | CAS | SELPNTEA | FFGQG |
| 22/1.1 | K2 | CAS | RDPDGVTEA | FFGQG | |
| 1488 | 22/1.1 | K1 | CAS | SMDHNTEA | FFGQG |
| 22/1.1 | K4 | CAS | KPDIGTEA | FFGQG | |
| 1276 | 22/2.5 | K1 | CAS | TQ | YFGPGTRLLV |
| 22/2.5 | K2 | CAS | TQ | YFGPGTRLLV | |
| 1376 | 22/2.5 | K2 | CAR | SGRKETQ | YFGPGTRLLV |
| 22/2.5 | K5 | CAR | SERKETQ | YFGPGTRLLV | |
| 1488 | 22/2.5 | K1 | CAS | SDNGRDIETQ | YFGPGTRLLV |
| 22/2.5 | K3 | CAS | SDNRDIETQ | YFGPGTRLLV |
- Amino acid sequences of the antigen-recognition sites of cloned PCR products specific for individual Vβ/Jβ combinations in three patients with autoimmune hepatitis (Patients 1276, 1376 and 1488). Sequence homologies within the N–D–N regions between clones from different patients are highlighted.
Clonal T-cell expansion was further confirmed with the sequence data from Vβ14/Jβ2.1- and Vβ14/Jβ2.2-expressing PCR clones from Patient 1276. Characteristic CDR3 motifs present in liver-infiltrating T cells were also found with some PCR products from peripheral blood T cells (Table 3).
| Vβ/Jβ | Origin | Clone | Vβ | N–D–N | Jβ |
|---|---|---|---|---|---|
| Vβ14/Jβ2.1 | Liver | K1 | CAS | SLSRGLLNEQ | FFGPGTRLTVL |
| Liver | K2 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Liver | K3 | CAS | SSSSSQ | FFGPGTRLTVL | |
| Liver | K4 | CAS | SSSSSQ | FFGPGTRLTVL | |
| Liver | K5 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Liver | K9 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Liver | K12 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Blood | K7 | CAS | SLSGGLLNEQ | FFGPGTRLTVL | |
| Blood | K8 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Blood | K10 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Blood | K11 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Blood | K12 | CAS | SLSRGLLNEQ | FFGPGTRLTVL | |
| Vβ14/Jβ2.2 | Liver | K2 | CAS | TRAAQYTGEL | FFGEGSRLT |
| Liver | K3 | CAS | TRAAQYTGEL | FFGEGSRLT | |
| Liver | K4 | CAS | TRAAQYTGEL | FFGEGSRLT | |
| Liver | K6 | CAS | TRAAQYTGEL | FFGEGSRLT | |
| Blood | K4 | CAS | SGRPSNTGEL | FFGEGSKLT | |
| Blood | K5 | CAS | SKTSGGVRGEL | FFGEGSRLT | |
| Blood | K6 | CAS | TRAAQYTGEL | FFGEGSRLT | |
| Blood | K8 | CAS | TERLGNTGEL | FFGEGSRLT |
- Amino acid sequences within the antigen-recognition sites of cloned PCR products from Patient 1276 expressing Vβ14, together with Jβ2.1 or Jβ2.2 molecules. Sequence homologies in the N–D–N regions between liver- and blood-derived PCR transcripts are highlighted.
Discussion
AIH is characterized by dense lymphocytic liver infiltrations and heterogenous serum autoantibodies. The specific target antigens for autoreactive T cells are unknown. Therefore, the aim of this study was to analyse whether disease-specific characteristic accumulation of T-cell clones was detectable in the liver-infiltrating and peripheral T cells. We analysed the T cells from three previously untreated patients with newly diagnosed type I AIH – a rare clinically observed condition. RT-PCR analysis and densitometric quantitation of PCR products revealed overexpression of multiple TCR Vβ-chains in the livers compared with that in the peripheral blood, as it was recently observed [14, 20]. To further characterize T cells that may accumulate at the inflammation site, CDR3 size spectratyping and PCR plot fragment analysis were performed. Multiple distinct single or twin peaks were found in AIH patients, suggesting common or individual clonal T-cell expansions with distinct CDR3 lengths [17, 21]. The nested PCR was conducted with selected PCR products of interest to further examine the usage of Jβ elements in combination with overexpressed Vβ elements. These experiments revealed that certain Vβ/Jβ combinations were strongly over-represented and common between AIH patients. Some overexpressed Vβ/Jβ combinations, however, were observed only in individual patients. To confirm the clonality of the expanded T cells, the CDR3 regions of PCR products that contained overexpressed usage of Vβ/Jβ elements were sequenced. The provided data clearly showed that many cloned PCR products with the same Vβ/Jβ combinations derived from the liver of an individual patient shared identical N–D–N diversity regions, indicating clonal T-cell expansions in the liver of AIH patients. In Patient 1276, identical CDR3 motifs were shared between Vβ14/Jβ2.1- and Vβ14/Jβ2.2-specific transcripts from liver-infiltrating and peripheral blood T cells; therefore, one could speculate that single expanded T-cell clones migrated to the liver tissue and were accumulated after they had been stimulated by unknown autoantigens.
Some amino acid sequences were found at characteristic positions of the N–D–N regions in different AIH patients, indicating disease-specific T-cell activation. This was also observed with in vitro-expanded T cells from patients with multiple sclerosis, which were specific to an immunodominant epitope and shared characteristic antigen-recognition sites [22]. T cells that recognized influenza or hepatitis C virus antigens showed predominant usage of certain Vβ/Jβ gene segments with common CDR3 motifs between individuals [23, 24]. Similarly, in type II AIH, the molecular analysis of in vitro-generated T-cell clones specific to a peptide from the LKM-1 antigen revealed common CDR3 motifs [15]. In this study, no identical CDR3 motifs were observed between cloned PCR products derived from different individuals, but the similarities of the antigen-recognition sites could indicate that in early type I AIH, oligoclonal T-cell infiltrates are directed against few immunodominant epitopes. Common CDR3 motifs have not been demonstrated in nine Japanese HLA-DR4-positive patients [2]. Thus, in later stages of AIH, antigen spreading may recruit further T-cell populations to the inflammation site [25]. Furthermore, discrepancies in the CDR3 motifs could reflect the fact that besides autoreactive T cells, not disease specific, bystander T cells also infiltrate the liver [11, 21].
The genetic background may influence T-cell recruitment to the inflammation site. In HLA-B27-restricted reactive arthritis, in vitro-expanded T-cell clones bore common Vβ/Jβ elements with identical CDR3 motifs [26]. Although frequently observed, type I AIH is not exclusively restricted to the DR3 or DR4 haplotype. These data may indicate that the diversity of CDR3 motifs could not be explained exclusively by the binding of different peptides by HLA class II molecules. These findings might have some clinical implication for therapeutic immune intervention. DNA vaccination in well-characterized experimental models with defined antigenic structures and genetic backgrounds deleted or suppressed disease-specific autoreactive T-cell clones [27]. However, in AIH, further studies with extended numbers of liver biopsy samples from patients with inflammatory active disease are needed.
In conclusion, T-cell clonotypes infiltrate the livers of patients with AIH after they have been stimulated by putative autoantigens. Besides disease-specific autoreactive T-cell clones, bystander T cells might also accumulate at the inflammation site. The similarities in the CDR3 motifs between AIH patients suggested few immunodominant epitopes that were relevant in the complex immunopathogenesis.
Acknowledgments
This paper was supported by the Deutsche Forschungsgemeinschaft (SFB 548-B3 and SFB 490-A4).




