The DNA binding and pairing preferences of the archaeal RadA protein demonstrate a universal characteristic of DNA strand exchange proteins
Abstract
The archaeal RadA protein is a homologue of the Escherichia coli RecA and Saccharomyces cerevisiae Rad51 proteins and possesses the same biochemical activities. Here, using in vitro selection, we show that the Sulfolobus solfataricus RadA protein displays the same preference as its homologues for binding to DNA sequences that are rich in G residues, and under-represented in A and C residues. The RadA protein also displays enhanced pairing activity with these in vitro-selected sequences. These parallels between the archaeal, eukaryal and bacterial proteins further extend the universal characteristics of DNA strand exchange proteins.
Introduction
The process of homologous recombination is an important pathway for generating genetic diversity, as well as for repairing damage resulting from DNA breaks. This fact has been augmented by the discovery of DNA strand exchange proteins throughout the Bacteria, as well as the Eukarya and the Archaea. These proteins are able to bring together two homologous DNA molecules and catalyse the exchange of their DNA strands, the central step in homologous recombination (Kowalczykowski et al., 1994; Bianco et al., 1998). The biochemical activities of DNA strand exchange proteins support their role in this pathway: they can assemble on single-stranded DNA (ssDNA) to form a helical nucleoprotein filament on ssDNA, hydrolyse ATP in a ssDNA-dependent manner and promote DNA strand exchange between homologous DNA molecules in vitro (Kowalczykowski, 1991; Sung, 1994; Seitz et al., 1998). Generally, binding to and exchanging DNA strands by these proteins has been considered to occur without regard to DNA sequence content. However, the fact that certain loci within prokaryotic and eukaryotic genomic DNA are enhanced or depressed for homologous recombinational activity raises the possibility that DNA strand exchange proteins harbour some DNA sequence preferences. Previously, it was shown that both the prototypic bacterial DNA strand exchange protein, RecA protein from Escherichia coli, and its eukaryotic counterpart, the Rad51 protein from Saccharomyces cerevisiae, show a preference for binding to certain GT-rich DNA sequences (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a). The specifically selected sequences also display an enhanced RecA or Rad51 protein-dependent pairing, suggesting that such GT-rich sequences are recombinationally more active. The sequences selected by these DNA strand exchange proteins also display similarity to known recombination hot-spots in bacteria, such as the E. coli recombinational hot-spot Chi (Stahl et al., 1975; Smith et al., 1980), and to genetic loci in eukaryotes that are known to display increased recombinational activity, such as microsatellite DNA (Jeffreys et al., 1998a, b), Alu repetitive elements (Marshall et al., 1996), triplet repeats from humans (Mariappan et al., 1999; Moore et al., 1999) and telomeric repeat sequences (Pryde and Louis, 1997; Stavenhagen and Zakian, 1998; Greider, 1999; Griffith et al., 1999). The fact that this preference for GT-rich DNA sequences was seen with the bacterial RecA protein and the eukaryal counterpart, Rad51 protein, prompted the question as to whether the same preference would be seen with the homologue from the third domain of life, the Archaea.
The archaeal RadA protein was discovered based on its homology to both the RecA and the Rad51 proteins, although this protein shows greater homology to the Rad51 protein (Sandler et al., 1996). RadA protein also exhibits similar biochemical characteristics to those of the E. coli RecA and S. cerevisiae Rad51 proteins, including ssDNA binding, nucleoprotein filament formation on ssDNA and DNA strand exchange activities (Seitz et al., 1998). However, the low ATP turnover rate and limited efficiency of DNA strand exchange exhibited by RadA protein suggests that this archaeal protein is more similar to the eukaryal Rad51 protein. As these three DNA strand exchange proteins originate from highly divergent organisms (Brown and Doolittle, 1995), we wanted to investigate whether the sequence preference displayed for both RecA and Rad51 proteins is also seen for the archaeal RadA protein. If this were the case, such a sequence preference could define a universal characteristic of RecA-like DNA strand exchange proteins.
Results
We used an in vitro selection experiment to determine whether the RadA protein displays a preference for DNA sequences with a particular composition. Starting with a pool of 6 × 1013 54-mers (SKBT18), composed of a random internal region of 18 nucleotides and two flanking defined regions of 18 nucleotides (Tracy and Kowalczykowski, 1996), we added limiting concentrations of RadA protein (to guarantee equal competition between all the 54-mers) under conditions in which the RadA protein would bind efficiently to the oligonucleotides. Five consecutive rounds of RadA selection and polymerase chain reaction (PCR) amplification were performed. Then, the selected 54-mers were cloned, and randomly chosen clones were sequenced. Table 1 shows the sequences of 40 internal 18-mer regions selected by the RadA protein. The average base composition of these sequences is highly over-represented for G residues (44.9%), although under-represented for A and C residues (9.2 and 18.3% respectively). The average base composition for T residues, however, remains slightly above the statistical average at 27.6%. One of the most interesting and intriguing results was that the RadA protein selected a sequence (the sequence displayed for clone 1) five times that was also selected by the RecA and S. cerevisiae Rad51 proteins out of this initial pool of 6 × 1013 oligonucleotides (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a). Additionally, the RadA protein selected a sequence four times (clone 6) that differed from the sequence of clone 1 by only a single base change.
| 1. | GCGTGTGTGGTGGTGTGC | |
| 6. | GGGTGTGTGGTGGTGTGC | |
| 10. | GGCTGTGTGGTGGTGTGC | |
| 11. | GGGACATATTGGTCGCTG | |
| 12. | GAACCTAGCATCTGTCCC | |
| 13. | AGAGAGCGGGTGTGGTCC | |
| 14. | GGAGGCTCCTTGCTTGTG | |
| 15. | ATCCTGCAGGTAGTGTGG | |
| 16. | GGATCCGTGTACACGTCC | |
| 17. | GGGTTAGGTATGTGTGCC | |
| 18. | GGGGGTGACTTACTGGCC | |
| 19. | GGGGGTTATGTTGGCCCG | |
| 20. | GGGGGATTGTGCGGTGGC | |
| 21. | GGGGAATTATACGTGGGG | |
| 22. | GGGTAGTTGGGGTTGTGC | |
| 23. | GTGGTGTGGTGCTGTGCC | |
| 24. | GCAGTGCTAGGGTAGGCA | |
| 25. | GGGATTATCCGCGTGTCT | |
| 26. | GGCGACATTACCCGTCCC | |
| 27. | GGGGTTAGAACTCGTGTC | |
| 28. | ACCAGGACGCTACCCGCC | |
| 29. | GCGATAGAGCTGTGTGCC | |
| 30. | GACCTGGTGCCTGTACCG | |
| 31. | GGGGTATGCACTTGGCCC | |
| 32. | GGGGCGGGTTACGTGTCC | |
| 33. | GGATGTTGCGTTGTTGTG | |
| 34. | GGCCGGTGACTTGTTGGC | |
| 35. | GATATGGTACACTGTCTG | |
| 36. | GGCCGGTGACTTGTTGGC | |
| 37. | GGGGATTGTATCGCTGTG | |
| 38. | GGGGAGTTGCAACTTGGC | |
| 39. | GGGGGTGGTTCGACTGCC | |
| 40. | GCGGTATAACATGCTGTC | |
| Totals | ||
| %A | 9.2 | |
| %C | 18.3 | |
| %G | 44.9 | |
| %T | 27.6 | |
- RadA protein selects for GT-rich DNA sequences from a random oligonucleotide pool. The sequences of the 18-nucleotide region are shown after five rounds of selection and amplification, together with the average base composition for all the sequences that were selected. Sequence 1 was obtained five times, and sequence 6 was obtained four times; the averages indicated reflect the multiple occurrences of these sequences.
As for RecA and Rad51 proteins, the RadA protein binds two or three nucleotides per protein monomer (Seitz et al., 1998). Therefore, we looked at the dinucleotide and trinucleotide distribution in the sequences selected by the RadA protein. Table 2 shows that there is a group of trinucleotides that is significantly over-represented in these sequences (GTG, TGT, GGT, GGG, TGG). These same five trinucleotides are over-represented in sequences selected by the S. cerevisiae Rad51 protein, and four of these five (GTG, TGG, GGT, TGT) are significantly over-represented in sequences selected by the E. coli RecA protein (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a). Additionally, many trinucleotides containing A and C residues are significantly under-represented, which is consistent with the results for RecA and S. cerevisiae Rad51 proteins.
| Trinucleotide | Frequency (%) | |
|---|---|---|
| GTG | 13.9 | |
| TGT | 9.1 | |
| GGT | 7.3 | |
| GGG | 6.4 | |
| TGG | 5.6 | |
| TGC | 3.8 | |
| TTG | 3.0 | |
| GGC | 2.5 | |
| GTT | 2.3 | |
| CTG | 2.0 | |
| CGT | 2.0 | |
| GCC | 1.9 | |
| GCG | 1.9 | |
| GTC | 1.6 | |
| GGA | 1.6 | |
| GCT | 1.4 | |
| ACT | 1.4 | |
| TCC | 1.4 | |
| CCG | 1.4 | |
| GAC | 1.3 | |
| TAG | 1.3 | |
| TTA | 1.3 | |
| TAT | 1.3 | |
| TAC | 1.3 | |
| CTT | 1.3 | |
| GTA | 1.3 | |
| GAT | 1.1 | |
| ACC | 1.1 | |
| CCC | 1.1 | |
| GCA | 0.9 | |
| AGG | 0.9 | |
| ATT | 0.9 | |
| CGG | 0.9 | |
| GAG | 0.8 | |
| ATG | 0.8 | |
| ATA | 0.8 | |
| CCT | 0.8 | |
| CGC | 0.8 | |
| ACG | 0.6 | |
| AGA | 0.6 | |
| ATC | 0.6 | |
| TCG | 0.6 | |
| TCT | 0.6 | |
| ACA | 0.6 | |
| AGT | 0.5 | |
| AGC | 0.5 | |
| AAC | 0.5 | |
| TGA | 0.5 | |
| CGA | 0.5 | |
| CTA | 0.5 | |
| CAT | 0.5 | |
| CAC | 0.5 | |
| AAT | 0.3 | |
| TTC | 0.3 | |
| CAG | 0.3 | |
| CAA | 0.3 | |
| CTC | 0.3 | |
| TTT | 0.2 | |
| CCA | 0.2 | |
| TCA | 0.0 | |
| AAG | 0.0 | |
| TAA | 0.0 | |
| GAA | 0.0 | |
| AAA | 0.0 |
- Trinucleotide frequencies contained in the sequences selected by the RadA protein. The frequency was determined as the number of times a certain trinucleotide occurred in all the sequences and divided by the total number of trinucleotides in the selected sequences. The expected statistical frequency is 1.6% (1/64).
Analysing the dinucleotide frequencies contained in the selected sequences reveals (Table 3) that three dinucleotides are significantly over-represented in the sequences selected by the RadA protein (TG, GT, GG). Again, these three dinucleotides were significantly over-represented in sequences selected by the S. cerevisiae Rad51 protein (Tracy et al., 1997a) and RecA protein (Tracy and Kowalczykowski, 1996). These results show that the RadA protein preferentially binds to DNA sites that are rich in G and T residues, compared with those containing A and C residues.
| Dinucleotides | Frequency (%) | |
|---|---|---|
| TG | 18.5 | |
| GT | 17.9 | |
| GG | 16.9 | |
| GC | 8.1 | |
| CC | 5.1 | |
| TT | 4.4 | |
| CG | 4.3 | |
| CT | 4.0 | |
| TA | 3.5 | |
| AC | 3.5 | |
| GA | 3.4 | |
| AT | 2.9 | |
| TC | 2.6 | |
| AG | 2.4 | |
| CA | 1.6 | |
| AA | 0.7 |
- Dinucleotide frequencies contained in the sequences selected by RadA protein. The frequency was determined as the number of times a certain dinucleotide occurred in all the sequences divided by the total number of dinucleotides in the selected sequences. The expected statistical frequency is 6.3% (1/16).
The RadA protein can promote homologous DNA pairing by mediating the invasion of supercoiled DNA by a homologous ssDNA molecule (Seitz et al., 1998). We therefore used this strand invasion assay to investigate the preference for joint molecule formation catalysed by RadA protein, with respect to sequence content. Previously, it was shown that both the RecA and the Rad51 proteins showed an enhanced rate and extent of joint molecule formation with an oligonucleotide containing selected sequence 1 (SKBT16) (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a). We tested RadA protein using oligonucleotides that contained the selected sequence (SKBT16), its complement (SKBT17), which pairs in the same place on the plasmid, or two other sequences (SKBT19 and SKBT20), which are homologous to the supercoiled DNA in a region outside which the selected sequences pair. Figure 1A shows that, under optimal conditions, RadA protein shows a more efficient homologous pairing reaction with selected sequence 1 (SKBT16) than with any other oligonucleotide; this is the same result as seen for both RecA and Rad51 proteins (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a). RadA protein pairs the selected DNA to a threefold greater extent (20%) than with the control DNA (5–8%), and it also pairs DNA containing selected sequence 1 (3.4% min−1) faster than the other DNA sequences (2.6% min−1).
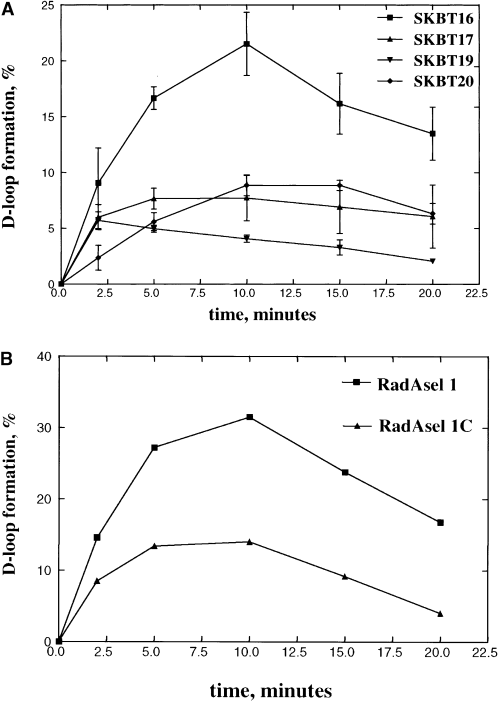
A. RadA protein preferentially promotes joint molecule formation with selected DNA. Joint molecule formation promoted by RadA protein was at 20 mM Mg2+ (optimal conditions) with pBT54CN1 and the following oligonucleotides (at 65°C): SKBT16 (▪, contains selected sequence 1); SKBT17 (▴, contains the complement of SKBT16); SKBT19 (▾, pairs on the opposite side of the plasmid); and SKBT20 (◆, pairs adjacent to SKBT16).
B. Reactions were carried out as above with a pUC19 supercoiled plasmid containing homology to the following oligonucleotides: RadAsel 1 (▪, contains the selected sequence) and RadAsel 1C (▴, contains the complement of this selected sequence).
To investigate this pairing preference further, we also performed the same joint molecule formation assays with another selected sequence, one that was uniquely selected by RadA protein (RadAsel 1: 5′-ATCCTGCAGGTAGTGTGG-3′). Figure 1B shows that pairing of this DNA by RadA protein displays about a threefold greater yield of joint molecules (30%) compared with the complement of this sequence (12%); furthermore, the rate for pairing the selected sequence (4.5% min−1) is greater than that for pairing the complement of this sequence (3.7% min−1). These results demonstrate that the RadA protein not only preferentially selects DNA that is GT rich, but it also homologously pairs that DNA more efficiently.
Discussion
The results presented here provide evidence that the archaeal DNA strand exchange protein, RadA, preferentially binds to GT-rich DNA sequences. This preferred binding is consistent with existing data for E. coli RecA, S. cerevisiae Rad51 and Homo sapiens Rad51 proteins (E. M. Seitz and S. C. Kowalczykowski, submitted), thus showing that this property is universal throughout all domains of life (Tracy and Kowalczykowski, 1996; Tracy et al., 1997b). Table 4 shows the frequency of the top five trinucleotides in the sequences selected by all four DNA strand exchange proteins examined to date. The most frequently selected trinucleotides are conserved among all four proteins, with the exception of GGG, which does not appear in the top five trinucleotides selected by the RecA protein. In this regard, the trinucleotides selected by the eukaryal and archaeal proteins are more similar, which is reasonable because of the closer evolutionary ties that the Archaea share with the Eukarya, compared with the Bacteria.
| RadAa(S. solfataricus) | Rad51b(S. cerevisiae) | Rad51c(H. sapiens) | RecAd(E. coli) |
|---|---|---|---|
| GTG | GTG | GTG | GTG |
| (13.9) | (10.4) | (17.3) | (7.6) |
| TGT | TGT | GGG | TGG |
| (9.1) | (7.4) | (12.8) | (7.6) |
| GGT | TGG | TGT | GTT |
| (7.3) | (7.2) | (9.3) | (6.3) |
| GGG | GGT | GGT | GGT |
| (6.4) | (7.0) | (7.8) | (6.0) |
| TGG | GGG | TGG | TGT |
| (5.6) | (5.6) | (6.6) | (6.0) |
- a . This work.
- b . Tracy et al. (1997) .
- c . E. M. Seitz and S. C. Kowalczykowski, submitted.
- d . Tracy and Kowalczykowski (1996).
- DNA strand exchange proteins select for DNA sequences that are primarily G-rich and under-represented for A and C residues. The five most frequently occurring trinucleotides found in all DNA strand exchange proteins examined are shown: E. coli RecA, S. cerevisiae Rad51, H. sapiens Rad51 and S. solfataricus RadA proteins, displaying a bias for trinucleotides exclusively composed of G and T residues.
For all DNA strand exchange proteins examined, the selected DNA sequences also showed an enhanced rate and extent of pairing to a homologous supercoiled plasmid compared with other control DNA sequences, but with no change in ATP hydrolysis activity (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a). Although the enhanced affinity of these complexes formed by the DNA strand exchange proteins for GT-rich sequences could explain the increased rate of joint molecule formation, this is unlikely to be the main reason, as the rate-limiting step in joint molecule formation is the opening of the recipient double-stranded DNA (dsDNA; Holloman and Radding, 1976). Rather, the enhanced pairing of DNA strand exchange proteins at these sequences probably results from a unique structure (although not a triplex structure) present in the selected dsDNA that facilitates the strand invasion step (R. B. Tracy and S. C. Kowalczykowski, unpublished).
No correlation between the GT richness of these selected sequences and the overall genome content of the various organisms exists. The GC content for each genome is as follows: E. coli– 51.5%; S. cerevisiae– 39.7%; Sulfolobus solfataricus– 36.5%. The absence of a correlation implies that the preference for GT-rich sequences shown by these proteins evolved independently of genome content. Instead, this preference must have some other fundamental basis. Recent data show that these GT-rich sequences display enhanced joint molecule formation even in the absence of any DNA strand exchange proteins (R. B. Tracy and S. C. Kowalczykowski, unpublished). The conclusion that these selected GT-rich sequences are intrinsically more receptive to homologous pairing leads to the proposal that they represent DNA sequences that were used for protein-independent pairing of DNA before the evolution of DNA strand exchange proteins. At a time when the DNA replication machinery required recombination to initiate replication or to elongate the DNA, these recombinationally active DNA sequences could facilitate DNA repair more easily by a recombinational mechanism (Kowalczykowski, 2000). It is therefore reasonable to speculate that DNA strand exchange proteins took advantage of this intrinsic property of GT-rich DNA to enhance the rate of homologous pairing. Such a scenario is consistent with the present-day importance of DNA recombination for the repair of double-strand breaks occurring during DNA replication (Morrical and Alberts, 1990; Kogoma, 1997; Michel et al., 1997).
The set of GT-rich sequences selected by all four DNA strand exchange proteins is similar to genetically unstable DNA sequences that exist in Bacteria and Eukarya (Tracy et al., 1997a), and are also GT rich (evidence for recombination hot-spots in archaeal genomes has not yet been uncovered). One striking similarity of these selected sequences was to the E. coli recombination hot-spot, Chi (5′-GCTGGTGG-3′) (Stahl et al., 1975; Smith et al., 1980), which is a DNA element important in regulating the nuclease activities of the RecBCD enzyme, as well as in RecBCD-dependent loading of the RecA protein onto a newly processed ssDNA molecule (Anderson and Kowalczykowski, 1997a, b). Not only is Chi a specific recombination hot-spot, but it is also embedded in DNA loci with a high G content and a low A content (Tracy et al., 1997b). Examples of recombinationally active sequences in eukaryotic organisms, to which the selected sequences are similar, include microsatellite DNA sequences (Jeffreys et al., 1998a, b), Alu repetitive elements (Marshall et al., 1996) and telomeric repeat sequences (Pryde and Louis, 1997; Stavenhagen and Zakian, 1998; Greider, 1999; Griffith et al., 1999). The incidence of GT-rich DNA at these genetically unstable regions and the preference for DNA strand exchange proteins to bind GT-rich DNA sequences may suggest that these proteins are important in these processes. This is further supported by recent evidence showing that the human Rad51 protein shows the same preference for GT-rich DNA (E. M. Seitz and S. C. Kowalczykowski, submitted).
The evidence presented in this paper, which demonstrates that the archaeal RadA protein has the same DNA binding and pairing preference as RecA and Rad51 proteins, clearly indicates that this is a universal characteristic of DNA strand exchange proteins.
Experimental procedures
In vitro selection assay
In vitro selection was carried out as described previously (Tracy and Kowalczykowski, 1996), except that the selection was initiated by the addition of 1 µM RadA protein and incubated at 65°C for 1 h before applying the reaction to the filter. After the five cycles were completed, the selected sequences were cloned into the plasmid pUC19. Transformants were selected and sequenced by the dideoxy-chain termination method using [α-32P]-dATP. Alkaline lysis and denaturation was used to prepare the DNA for sequencing (Tracy and Kowalczykowski, 1996; Tracy et al., 1997a).
Joint molecule formation assays
Oligonucleotides were 5′ end-labelled with [γ-32P]-ATP and T4 polynucleotide kinase. Joint molecule formation took place in a reaction mixture (260 µl) with 25 mM Tris acetate, pH 7.5, 20 mM Mg acetate, 1 mM dithiothreitol (DTT), 2.5 mM ATP, 1 µM nucleotides γ-32P-labelled 54-mer oligonucleotide, 0.33 µM RadA protein, 18 µM nucleotides plasmid DNA (pBT54CN1, a pUC19 derivative; Tracy and Kowalczykowski, 1996). Reactions were initiated by the addition of RadA protein after preincubation of the other components for 5 min at 65°C. At 0, 2, 5, 10, 15 and 20 min, 40 µl aliquots were removed and added to 5 µl of a 10% SDS−0.5 M EDTA mixture and 5 µl of DNA loading buffer. The reactions were subjected to agarose gel electrophoresis in 1% agarose gels run in TAE buffer for 180 V h. The gels were dried on DE-81 paper, imaged using a Storm phosphorimager and quantified using imagequant 4.0 software. The percentage of joint molecule formation was determined relative to the limiting amount of plasmid DNA used in each reaction.
Acknowledgements
We would like to thank the following members of the Kowalczykowski laboratory for comments on this manuscript: Rick Ando, Piero Bianco, Frank Harmon, Julie Kleiman, Alex Mazin, Susan Shetterly, and special thanks to Jim New and Frederic Chedin for extra help reviewing this manuscript. The work described in this manuscript was supported by NIH grant AI-18987 and HFSP-RG63 to S.C.K. and by NIH training grant GM07377 to E.M.S.