Rangewide variation of the maritime pine bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) in relation to the genetic structure of its host
Abstract
The bast scale Matsucoccus feytaudi is a specific pest of maritime pine, but the damage inflicted by the insect on the host trees is variable, ranging from no apparent effect to severe decline of the maritime pine stands. Rangewide variation of mitochondrial DNA among M. feytaudi populations was analysed by polymerase chain reaction–restriction fragment length–single-strand conformation polymorphism (PCR–RFLP–SSCP) analysis and the results compared with the genetic information already available for its host. Three main nonoverlapping lineages can be distinguished in M. feytaudi. The phylogeography of the pest population is clearly related to the history of its host. Most local associations could result from common evolution while others must be interpreted as intraspecific host shifts. Because the distribution of cultivated tree species is greatly influenced by humans, much may be learned concerning their genetic structure from the indirect study of their specific pests.
Introduction
The bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) is a specific pest of maritime pine (Pinus pinaster Ait.). The distribution of the insect is therefore included within the range of its host, which occurs as fragmented populations in the western part of the Mediterranean basin. Western populations of the insect are endemic whereas eastern populations are known to have been recently introduced. The insects caused severe outbreaks responsible for the decline of 120 000 ha of maritime pine in southeast France (Schvester 1967), and are presently spreading in the north of Italy (Covassi & Binazzi 1992). An epidemic evolution is also feared in Corsica where a bast scale population was discovered in 1994 (Jactel et al. 1996). The biology of the insect and the description of the damage generated under epidemic conditions are reviewed by Riom (1994).
Level of damage is assumed to be dependent on the environmental conditions, the presence of secondary pests and the differential susceptibility of the host tree (Schvester & Ugetto 1986; Carle 1973; Riom 1980). Harfouche et al. (1995) described the existence of a longitudinal gradient in the susceptibility of maritime pine to M. feytaudi, increasing from Morocco to Italy. They suggested that this gradient is related to the duration of exposure to the insect and consequently hypothesized a west-to-east dispersal of the pest, from Moroccan populations. While the intraspecific diversity of maritime pine is well documented, potential variability of the insect itself has never been explored. The purpose of this study is to present a survey of M. feytaudi genetic variation and to examine its relations with the genetic structure of its host. This would help to determine whether levels of resistance of the host tree have been evolutionarily adjusted.
The rangewide genetic structure of M. feytaudi populations was thus investigated by the analysis of mitochondrial DNA (mtDNA), a nonrecombinant haploid and maternally inherited marker extensively used in insect phylogeographic studies (Roderick 1996; Avise et al. 1987; Moritz et al. 1987; Avise 1998). These intrinsic features are responsible for its particular sensitivity to the effect of random drift (Birky et al. 1989).
The analysis of mtDNA PCR fragments was based on the single-strand conformation polymorphism technique (SSCP), a simple and sensitive technique for detecting polymorphisms affecting the conformation of single-stranded DNA in nondenaturing acrylamide gels (Black IV & Du Teau 1997). As this technique is more sensitive for small fragments (Hayashi 1991), and because the resulting information depends on the number of fragments analysed, the use of restriction enzymes prior to the SSCP analysis for the larger PCR fragments (Iwahana et al. 1992) is recommended (Dumolin-Lapègue et al. 1996). The different haplotypes detected by SSCP can be subsequently sequenced, providing appropriate data for phylogenetic analysis (Orti et al. 1997).
Materials and methods
Insect collection
The small size of the insect and the apparent absence of damage generated by endemic populations made detection and sampling difficult until the identification and synthesis of the sex pheromone allowed pheromone trapping of the males (Einhorn et al. 1990; Jactel et al. 1994). It is now possible to carry out a comprehensive sampling of the insect throughout its range. Traps were baited with lures loaded with 50 µg of synthetic pheromone (Jactel et al. 1994). One trap per locality was placed in maritime pine stands from February to May. Insects were collected twice a month and the lure renewed each month. Insects were stored in plastic boxes containing silica gel prior to being sorted. After sorting, dried Matsucoccus feytaudi were stored at −80 °C. Depending on the number of insects that were trapped (ranging from none to a few thousand individuals per locality per date) and the quality of the collection (15% of the specimens did not provide any amplification signal, probably due to DNA degradation before freezing), 3–22 individuals from 34 localities were used for genetic analyses. Insects collected from a single trap at different dates were considered to represent a population.
Experimental procedures
Extraction. Genomic DNA of single males was obtained using 30 µL of 5% chelex solution (Walsh et al. 1991) as described in Vanlerberghe-Masutti (1994).
Primers. Conserved regions greatly facilitate the design of primer pairs, allowing the amplification of mtDNA fragments from various taxa (Simon et al. 1994). Because scale insects have received little attention so far from molecular geneticists, it was necessary to screen among available primers those allowing the amplification of M. feytaudi mtDNA. More than 40 primers were tested in different combinations, mainly from the University of British Columbia Insect Mitochondrial DNA Primer Oligonucleotide Set provided by J. Hobbs and described in Loxdale & Lushai (1998). Most of the primer pairs tested failed to amplify M. feytaudi mtDNA. Succesful primers pairs are provided in Table 1.
| Primer pair | Corresponding genes amplified in Drosophila (size of the fragment) | Annealing temp. (°C) | Extension time (min) |
|---|---|---|---|
| C1-J-1718/C1-N-2191 | part of COI (473 bp) | 50 | 1 |
| CI-J-2195/C2-N-3661 | part of COI-tRNAleu-part of COII (1466 bp) | 52 | 2 |
| CB-J-10933/CB-N-11367 | part of Cyt b (434 bp) | 45 | 1 |
| SR-J-14233/SR-N-14588 | part of 12S rDNA (355 bp) | 55 | 2 |
PCR. PCR conditions were modified from Moreau et al. (1994). Amplification reactions were carried out in a total volume of 25 µL containing 16.6 mm (NH4)2SO4, 67 mm Tris-HCl pH 8.0, 2 mm MgCl2, 0.001% anionic detergent, 10 mmβ-mercaptoethanol, 4.4 µg/mL bovine serum albumin, 200 µm of each dATP, dGTP, dCTP, dTTP, 1 unit of Goldstar Polymerase (Eurogentec), 0.2 µm of each primer, and 2 µL of chelex supernatant. Temperature cycling was carried out in a PHC3 Techne PCR, starting with 4 min at 94 °C, followed by 30 cycles of 45 s at 92 °C, 45 s at the annealing temperature, 1 or 2 min at 72 °C and a final extension of 10 min at 72 °C (Table 1).
RFLP–SSCP. The SSCP technique was applied directly to the smallest fragments amplified (< 500 pb). For the larger fragment (COI–COII), prior digestion with the restriction enzymes HinfI and DraI was used. The SSCP technique (Orita et al. 1989) and the silver-staining procedure used (Bassam et al. 1991) are described in Bodénès et al. (1996).
Sequencing. Sequencing procedures were modified from Dumolin-Lapègue et al. (1998). New pairs of primers were synthesized by adding forward/reverse M13/pUC sequence at the 5′ end of SR-J-14233/SR-N-14588, respectively. PCR products were purified using the Qiaquick PCR purification kit (Qiagen). Sequencing reactions were performed with the dye primer cycle sequencing core kit (PE applied Biosystems), using M13/pUC forward and reverse labelled primer. Sequencing was carried out using a 4000 L automatic DNA sequencer (Li-Cor) using 6% Long Ranger (TEBU) gels.
Data analysis. Diversity and differentiation analysis followed methods described in Pons & Petit (1995, 1996), using the program haplodiv. Genetic differentiation was determined in two ways: either genetic similarities between haplotypes (proportion of shared fragments) were taken into consideration (NST) or only frequencies were used (GST). These two parameters can be compared using an analytical test (Pons & Petit 1996). We also designed a permutation test to confirm the difference: 1000 random permutations of haplotype identities were made, keeping the haplotype frequencies and the matrix of pairwise haplotype differences as in the original study. The distribution of values obtained by permutation was compared with the observed value. clustalw version 1.7 (Thompson et al. 1994) was used to determine sequence alignments and to construct phylogenetic trees based on the neighbour-joining method.
Results
Distribution of Matsucoccus feytaudi
Insects were caught by pheromone trapping in all regions where the presence of the insect had already been reported by direct observation (Riom 1980) (Fig. 1). However, the insect was recently found in one forest in Brittany, despite the negative results obtained during our study in three different forests of this region. M. feytaudi is present in most of the stands examined in the western part of the maritime pine natural range. Its absence in a few localities could be related to the non-native status of the maritime pine stands and to their distance from natural stands, for instance in the Maâmora forest in Morocco. The absence of the insect is also notable in the intermediate stands of Corbières (southeast France), a region where the autochthonous status of maritime pine is questionable (Baradat & Marpeau 1988). On the other hand, M. feytaudi reaches northern stands in Pays de Loire, where maritime pine is known to be an introduced species.
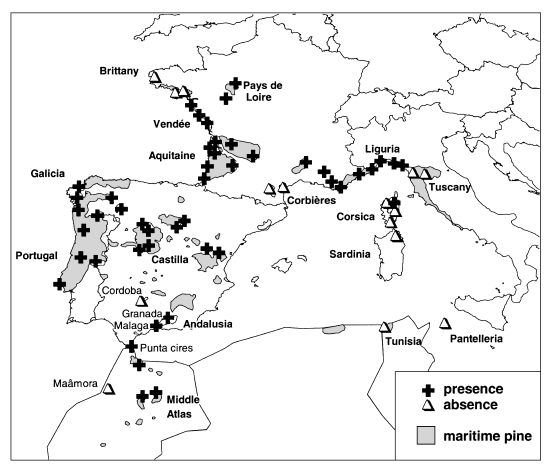
Natural range of Matsucoccus feyatudi established by peromone trapping.
The situation in the eastern part of the maritime pine natural range is directly related to the recent introduction of the insect in southeast France where it is now widely distributed. It has subsequently colonized the northern Italian stands of Liguria, and continues to spread towards Tuscany. In Corsica, only the northeastern stands are colonized at the present time. Maritime pine forests from Sardinia, Pantelleria and Tunisia are free of the insect.
Haplotypes
We obtain SSCP data from five or six mtDNA fragments, depending on the absence or presence of the HinfI restriction site in COI–COII. All fragments were polymorphic, and the combined RFLP–SSCP analysis of the four mtDNA fragments amplified allowed the detection of 22 different mitotypes (Table 2). The distribution of these haplotypes is illustrated in Fig. 2.
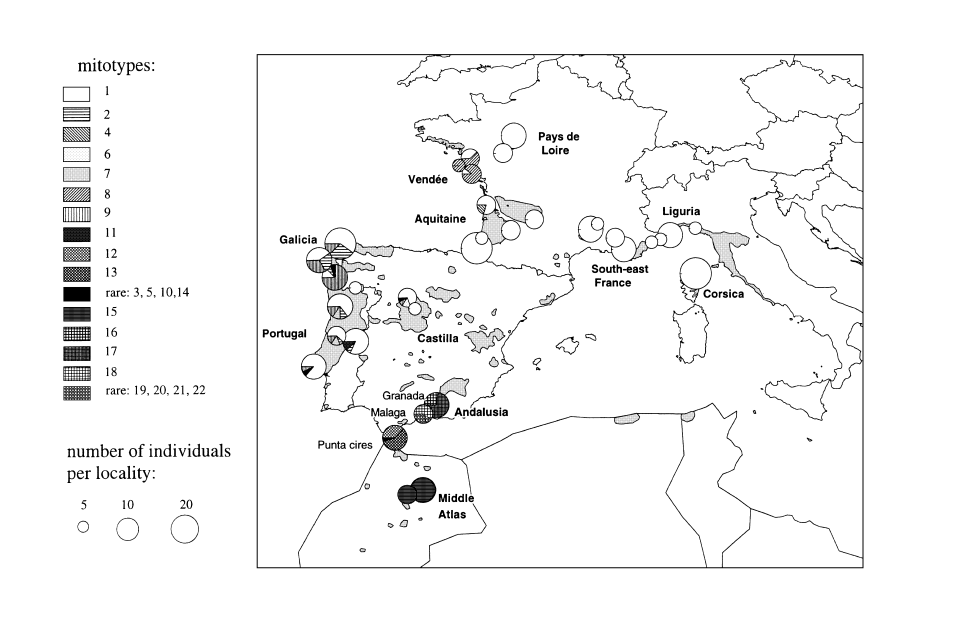
Distribution of the mitotypes of Matsucoccus feytaudi detected by PCR–RFLP–SSCP.
After purification, the direct sequencing of cyt-b, COI and COI–COII fragments failed, certainly because of the presence of additional fragments in the PCR products. Further investigations (using cloning, double PCR and the design of new primers) allowed us to sequence these fragments from other individuals (our unpublished data). The similarity among these sequences and those corresponding to the same mtDNA genes in other insects indicates that the expected fragments had been amplified in our survey.
For the fragment amplified using 12S rDNA primers, we obtained the complete sequence of the PCR product. Its size (330–332 bp) is smaller than the 355 bp of the corresponding fragment amplified in Drosophila. Similarity searches using the blast algorithm (Altschul et al. 1990) revealed no similarity among our sequences and the numerous sequences of the third domain of the 12S rDNA gene compiled in sequence databases. Low matches were found with different sequences, mainly from the mtDNA control region of various animals, due to their high A + T content. Efforts to search for a secondary structure similar to the skeleton of the 12S rDNA third domain proposed by Hickson et al. (1996) failed. Sequencing the PCR product obtained with the same primers in M. josephi (from Israel) resulted in a high level of homology.
Diversity and differentiation
Estimates of diversity and differentiation were based on the RFLP–SSCP data only. As the nature of the fragment amplified using 12S rDNA primers remains uncertain, the corresponding information was not used here. A low level of intrapopulation diversity (hS = 0.17) and a high level of total diversity (hT = 0.53) were found, indicating that the diversity is mostly distributed among populations (GST = 0.67). In actual fact, the situation is heterogeneous: eastern (southeast France and Italy) and southern (Middle Atlas, Morocco) populations are fixed for a single mitotype, while most of the Iberian populations (Spain and Portugal) are polymorphic.
To consider the level of divergence between haplotypes, we computed NST following Pons & Petit (1996). The matrix of distances between haplotypes was based on the number of fragments differing in their RFLP–SSCP patterns. The value obtained (NST = 0.87) is significantly higher than GST, based on the test proposed by Pons & Petit (1996) (U = 3.57, P < 0.01). This result was confirmed by permutation analysis of haplotype identities: the highest NST value obtained among the 1000 permutations was 0.76 (with a mean of 0.67, identical to the observed GST value). This maximum NST value is much lower than the observed one, indicating that it is important to take into account the similarities between haplotypes to measure how diversity is distributed among populations, probably a consequence of the existence of a phylogeographic organization of M. feytaudi mtDNA diversity.
The phylogenetic relationships between haplotypes are illustrated in Fig. 3. The SSCP patterns of the fragments amplified is weakly informative for phylogenetic purposes. However, three major lineages can be distinguished. A first lineage includes most western European populations and the Punta Cires population from Morocco, a second corresponds to the Andalusian populations and a third is represented by the only mitotype found in the Middle Atlas of Morocco. The second and third lineages share the absence of the HinfI restriction site in COI–COII. The tree inferred from the sequences of the fragment amplified with the 12S rDNA primers (Fig. 4) is consistent with these three lineages, with haplotypes from Andalusia and Middle Atlas sharing two substitutions. One can notice that among the nine substitutions observed, only one is homoplasious. Length variations in the mononucleotide stretch (polyA) differentiate haplotypes within lineages, but seems to be too homoplasious to be used as a phylogenetic character at the within-species level.

Relationships between haplotypes inferred by RFLP–SSCP analysis of Matsucoccus feytaudi mtDNA fragments.
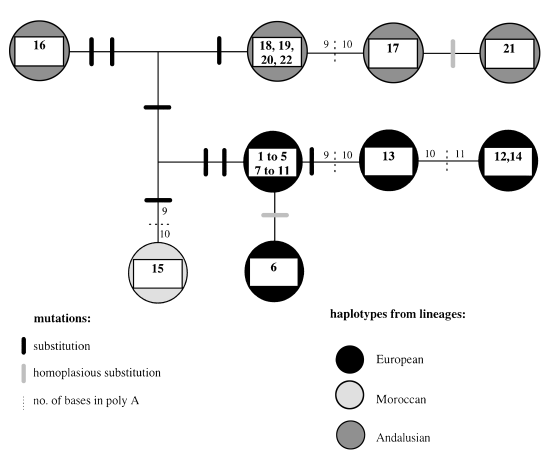
Phylogenetic tree inferred from the sequence of fragments amplified with primers correponding to 12S rDNA.
Discussion
PCR, combined with RFLP and SSCP, proved to be a powerful technique to investigate the rangewide variation of mtDNA in the scale insect Matsucoccus feytaudi. The haplotypes identified can be subsequently sequenced to fully identify their relationships (Stothard et al. 1998). Although the nature of the fragment amplified with 12S rDNA primers remains uncertain, the phylogenetic relationships based on its sequence are consistent with those obtained with the other fragments. Furthermore, a homologous sequence was obtained in the congeneric but geographically distant M. josephi. This fragment could therefore originate from another mtDNA region of M. feytaudi, rather than from its nuclear genome. We cannot rule out the possibility that we have amplified DNA from an endosymbiont, as strictly congruent phylogenies have been found between other homopteran species and their symbiotic bacteria (Douglas 1998).
The diversity and differentiation analysis uncovered a strong phylogeographic structure in M. feytaudi. This structure may be related to the complex history of its host tree. Hence, the comparison of the genetic structures of the insect and its host should improve our understanding of the evolution of each species (Pellmyr et al. 1998).
Genetic structure and gene flow
Climatic changes are known to have played a major role in shaping the phylogeography of European species by contracting and expanding their natural ranges (Hewitt 1996). By compiling several data sets available for various animals and plants, Taberlet et al. (1998) were able to identify some general trends for European species, according to the location of their refugia in southern Europe, but they concluded that each species will exhibit a particular phylogeographic pattern. Fossil records have identified the presence of P. praepinaster in Portugal three million years ago, from which modern maritime pine became differentiated at the end of the Pliocene (Teixeira 1945). Successive Quaternary ice ages have then moulded the range of the species, by isolating refugia which subsequently provided the source for new colonization. But the distribution of maritime pine has also been greatly modified by human activities during historic times all around the Mediterranean basin until the recent expansion of its cultivated range.
The fragmentation of the range of maritime pine, due notably to the presence of calcareous outcrops all around the Mediterranean basin, may have favoured the high degree of differentiation that we observed among M. feytaudi populations. Based on a suite of different nuclear markers, maritime pine was found to be characterized by a high coefficient of genetic differentiation among populations (GST = 0.17, Petit et al. 1995), when compared with other conifer species (mean of GST = 0.07, Hamrick et al. 1992). Insects characterized by strongly fragmented natural ranges are also known to have strongly differentiated populations (reviewed in Peterson & Denno 1998). Moreover, M. feytaudi mainly disseminates by a passive airborne transport of the first larval stage. The colonization of new areas depends on the presence of maritime pine trees that are at least 5-years old. The efficiency of such a dispersion process can be extrapolated from the annual advance of epidemic populations in newly infested stands, i.e. no more than 5–10 km each year. Natural dispersion of M. feytaudi is therefore likely to be strongly affected by the fragmented range of its host, as forests are often separated by tens or hundreds of kilometres, although rare long-distance migration events due to high winds cannot be excluded. Artificial infestation can also occur via transport of contaminated material, as proposed for the polemochore populations of M. josephi in Israel (i.e. dispersed in connection with war activities, Mendel et al. 1994).
Comparative phylogeography of the insect and its host: a shared history?
Three insect lineages (western European, Andalusian and Morrocan) have been identified on the basis of the PCR–SSCP analysis of the mtDNA fragments and can also be recognized in the phylogenetic tree inferred from the sequence of the fragment amplified with 12S rDNA primers. The genetic variation of maritime pine has been studied using various markers, such as terpenes (Baradat & Marpeau 1988), total proteins (Bahrman et al. 1994), isozymes (Petit et al. 1995) and chloroplast microsatellites (Vendramin et al. 1998). Some palynological and palaeoclimatological data are also available (reviewed in Baradat & Marpeau 1988). Most authors discriminate three major groups of populations, as defined by Baradat & Marpeau (1988). An Atlantic group comprises populations from western France, Portugal and a large part of Spain. The Mediterranean group consists of all eastern European populations, plus the Andalusian stands of Sierra Nevada and the small stand of Punta Cires in Morocco. The North African group includes populations from Pantelleria, Tunisia, Rif and Middle Atlas in Morocco plus the Andalusian stand of Sierra Ronda.
Western European lineage
Most of the scale populations characterized by the western European mtDNA lineage are associated with the Atlantic group of maritime pine. The geographical distribution of mtDNA diversity within this western lineage of M. feytaudi is compatible with an Iberian refugium for the insect and hence its host (Fig. 2). This confirms earlier hypotheses based on maritime pine studies (Baradat & Marpeau 1988), and is also supported by results on refugia and recolonization routes in western Europe for other tree species such as the white oaks (Dumolin-Lapègue et al. 1997), or the cork oak (M. P. Jimenez, L. Gil, R. J. Petit unpublished results).
The Iberian peninsula includes the largest part of the maritime pine natural range, where the stands are more or less contiguous, allowing migration of the insect and hence potentially reducing differentiation among populations. The relatively important mixing of mtDNA haplotypes in the western part of the range could be due to human influences, through planting or through the transport of contaminated logs. The remarkable extension of maritime pine forest in Portugal during the 20th century can explain both the absence of genetic structure for the tree, as revealed by chloroplast microsatellite markers (M. Ribeiro, personal communication), and a similarly blurred geographical pattern for M. feytaudi in this country.
The lower level of diversity found in western France could be related to a more recent establishment of the insect and/or genetic drift, as maritime pine was present mainly in a thin coastal range before its recent artificial expansion inland during the last two centuries. The presence of an endemic haplotype in Vendée, at the extreme north of the natural range of its host, could be due to the effect of drift in these small formerly isolated maritime pine stands. It also argues for a native status of maritime pine in Vendée, a hypothesis that could not be assessed with available palynological data.
The decline of eastern maritime pine stands, all belonging to the Mediterranean group, started in 1957 in two nearby forests on the French Mediterranean coast (Schvester et al. 1970) and then spread to northern Italy and Corsica. The absence of mtDNA variation in these regions is certainly related to the common origin and recent introduction of M. feytaudi, and can be interpreted as the result of a founder effect (Villablanca et al. 1998). The situation is similar in the artificial stands in Pays de Loire, where only the same haplotype was detected.
At the southern edge of the distribution of this western European lineage, the Moroccan Punta Cires population is characterized by the presence of endemic haplotypes, certainly due to its extreme and isolated geographical position. The genetic affinity of this population with the western European lineage is rather surprising, considering the greater geographical proximity of populations characterized by the divergent Andalusian and Moroccan mtDNA lineages. Moreover, although the maritime pine population of Punta Cires was considered by Baradat & Marpeau (1988) to be genetically marginal, it was nevertheless included in the Mediterranean group. Given the importance of commercial exchanges between the two continents, the native status of most forest tree populations and of their associated insects living in the vicinity of the Straits of Gibraltar is questionable.
Morrocan lineage
The high mtDNA divergence observed in the Middle Atlas for M. feytaudi is matched by the presence of a highly divergent maritime pine group. This may be interpreted as a result of their common history, Baradat & Marpeau (1988) arguing for a differentiation of the North African group during earlier ice ages. The absence of mtDNA variation in the two studied populations may be due to the action of drift in small isolated populations, but is also compatible with a recent founder effect. This latter scenario would be consistent with an introduction of maritime pine from the Rif to the Middle Atlas after the Arabian invasion during the 8th century, as Reille (1977) suggested on the basis of palynological data.
Andalusian lineage
The Andalusian populations exhibit a more complex pattern. All the haplotypes identified belong to a specific Andalusian mtDNA lineage, which is particularly divergent from the western European lineage. Contrary to the situation in the Middle Atlas, the level of polymorphism observed in Andalusia is remarkable, indicating that scale populations (and hence maritime pine forests) have remained important in this region. Baradat & Marpeau (1988) proposed two possible origins for the maritime pine stands of Sierra Ronda, which is the closest to the insect population sampled (near Malaga). A first hypothesis is an artificial introduction from Morocco (when the Omeyade dynasty arrived in Andalusia during the 8th century). Alternatively, an old endemism may be proposed. Although the Sierra Ronda pine population was included in the North African group, its divergence suggested a separate refugium during the last ice age. This interpretation is compatible with the high level of mtDNA divergence found for the Andalusian populations of M. feytaudi, and to its lower divergence from the Moroccan lineage than from the western European one.
However, although M. feytaudi individuals sampled near Granada are also characterized by this Andalusian mtDNA lineage, the maritime pine stands from the same region (Sierra Nevada) have been classified in the Mediterranean group. Baradat & Marpeau (1988) noted that Ionian colonies were present from 750 to 550 bc simultaneously in Andalusia and in southeast France. They may have introduced maritime pines from southeastern France, which could have been subsequently colonized by the local stock of endemic M. feytaudi populations. No damage has been registered in these stands, nor in other eastern Spanish stands also classified in the Mediterranean group. It would therefore be of great practical interest to compare these stands with the susceptible ones from southeastern France and Italy, which belong to the same maritime pine group, in order to see if the resistance has evolved de novo or through hybridization with local resistant trees belonging to the north African or Atlantic group.
Knowledge of population structure is often used to reconstruct the differentiation process of insect populations associated with different host species (Diehl & Bush 1984; Roderick 1996; Brown et al. 1997). The comparison of the phylogeography of the two protagonists, in the case of specific insect–plant associations, can also be of great interest, not only for the study of the insect but also for the evolution of its host, as shown here and in another recent investigation (Pellmyr et al. 1998). But palaeontological and historical data, especially in the case of a cultivated tree species, will remain essential.
Acknowledgements
We particularly thank T. Aumonier, M. Bertin, R. Bigel, C. Boyer, P.-Y. Caudal, H. Combeau, J.-M. Corti, N. Delaire, R. Delpont, V. Didier, S. Dubois, C. Erishmann, M.-R. Fleisch, X. Grenier, J.-F. Gueguen, R. Icher, M. Kleinhentz, J. Lecoq, J.-P. Lerol, G. Leroy, A. Lévy, J.-M. Linder, F. Mathieu, G. Martin, P. Ménassieu, J. Mirault, T. Noblecourt, S. Normand, J. Regad, C. Rulliere, F. Trottet, C. Vidal (France), Z. Mendel (Israel), A. Binazzi, M. Covassi, P. Luciano (Italy), D. Ghaioule (Morocco), M. T. Cabral, L. Nunes, J. C. Santos Silva, E. Sousa, T. Vasconcelos (Portugal), F. J. F. Ana Magan, P. Cabezuelo, J. S. Gutierrez, R. Hernandez Alonzo, M. J. Lombardero Diaz, M. Mozos Pascual, M. R. Ocón, J. M. Sierra Vigil (Spain), M. L. Ben Jamaa, K. Hamdi and M. Othmani (Tunisia) for help with the pheromone trapping. Sequence analyses were performed using the server INFOBIOGEN (Villejuif, France). The first author owes much to the technical and scientific assistance of C. Bodénès, S. Dumolin-Lapègue and R. Streiff.
References
This study is part of a research programme dealing with the evolutionary and functional approach of bast scale–pine relationships at the Laboratoire d’Entomologie Forestière, INRA Bordeaux, which is conducted by Hervé Jactel. Emmanuel Carcreff is a PhD student involved in this project. The work of Christian Burban focuses on the molecular evolution of insect populations and benefits from a close collaboration with Rémy Petit, a population geneticist interested in the phylogeography of forest trees at the Laboratoire de Génétique et Amélioration des Arbres Forestier, where the molecular investigations were carried out.




