Possible causes of morphological variation in an endemic Moroccan groundsel (Senecio leucanthemifolius var. casablancae): evidence from chloroplast DNA and random amplified polymorphic DNA markers
Abstract
Genetic variation was assessed in Senecio leucanthemifolius var. casablancae (Compositae), a Moroccan Atlantic coast endemic, in order to examine possible causes of atypical leaf morphology in three populations south of the known range. Evidence for introgression from S. glaucus ssp. coronopifolius and/or divergence was investigated with molecular markers. Both random amplified polymorphic DNA (RAPD) and chloroplast (cp) DNA restriction fragment length polymorphism (RFLP) differentiated the species well. Some evidence that hybridization may have occurred between the two species was provided by cpDNA markers. However, biparentally inherited RAPD markers failed to provide any support for the hypothesis that intermediate leaf morphologies in atypical populations arose through hybridization. Consequently, they are most likely to have arisen via divergence caused by drift and/or selection. Genetic distances among populations of S. leucanthemifolius were significant in all but one case. Isolation by distance was indicated by a significant positive correlation between genetic and geographical distances (r = 0.68, P = 0.01, Mantel test). These results suggest that long-distance achene dispersal is rare, despite the presence of a well-developed pappus. The observed loss of pappus at achene maturity may explain this unexpected result. Due to the morphological distinction of var. casablancae from other varieties of S. leucanthemifolius, we suggest elevation to species rank and treatment of the atypical material at infraspecific rank.
Introduction
Geographic variation in species may reflect population divergence due to insufficient gene flow to counteract the forces of drift and selection (Wright 1978; Slatkin 1985). Such divergence leads to the formation of geographical races and this has frequently been taken to represent the initial stages of speciation (Stebbins 1950; Clausen 1951; Grant 1981; but see Levin 2000). However, essentially similar patterns of variation can result from hybridization and introgression. Hybridization involving rare taxa has generally been viewed as a potential threat to survival (Avise 1994; but see Arnold 1997). Conservation concerns arise when introgression threatens to undermine the genetic integrity of a taxon (Brochmann 1984; Rieseberg et al. 1989; Rieseberg 1991; Rieseberg & Gerber 1995; Levin et al. 1996). On the other hand, introgressive hybridization can be a creative force, resulting in the evolution of new hybrid taxa on occasion (Abbott 1992; Arnold 1997; Rieseberg 1997).
We have recently noted a potentially interesting pattern of leaf-shape variation in the endemic Moroccan groundsel Senecio leucanthemifolius Poiret var. casablancae Alexander (Compositae). This pattern could either be the product of population divergence or the introgression of genes from the related S. glaucus L. ssp. coronopifolius (Maire) Alexander. We have employed molecular markers in an attempt to distinguish between these two possible causes of the morphological variation observed.
Senecio leucanthemifolius var. casablancae is a diploid (2n = 20) annual plant restricted to dune habitats along the Moroccan Atlantic coast. Bagged capitula in this taxon set virtually no seed (< 1%) (M. Coleman, pers. obs. 1999), indicating self-incompatibility. This result is consistent with previous work on the breeding systems of closely related diploid species (Alexander 1975; Abbott & Forbes 1993). Although not rare in numerical terms, this taxon is a localized endemic that may be regarded as vulnerable due to its restricted distribution and specialized ecology. The original description of the variety records a distribution between Salé in the north and Cap Beddouza in the south (∼300 km) (Alexander 1979). Senecio leucanthemifolius as a whole is widespread in the Mediterranean. Alexander (1979) divided the species into eight varieties, but one of these is generally treated as a distinct species S. vernalis Waldst. & Kit. The taxonomy of the species is complicated by intergradation between some varieties (Alexander 1979). However, S. leucanthemifolius var. casablancae is well differentiated from other varieties.
Three atypical populations displaying unusual leaf morphology have been discovered south of the known range of var. casablancae (M. Coleman & R.J. Abbott, pers. obs. 1999). Typical var. casablancae has oblong rhomboid rarely shallowly lobed leaves (Fig. 1A). This morphology appears to be stable across the originally described range. In contrast, the three southern populations each display distinct leaf morphologies: (i) variably lobed; (ii) coarsely toothed; and (iii) extremely slender and variably lobed (Fig. 1B–D). Leaf morphology is largely uniform within a given population, with the exception of the degree of lobing, but variable between populations as little as 30 km apart. Senecio is known to exhibit high levels of leaf-shape plasticity (Alexander 1975), but a strong genetic basis can be assumed in this case as the lobed and unlobed forms are maintained under cultivation. We refer to the atypical populations collectively as ‘atypical-casablancae’.
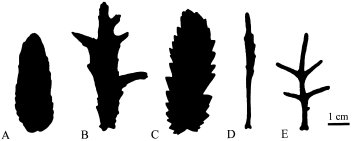
(A–D) Middle cauline leaf silhouettes of Moroccan Senecio. A, S. leucanthemifolius var. casablancae; B, ‘atypical-casablancae’ from Souira Kédima (SK); C, ‘atypical-casablancae’ from El-Mehattat (ME); D, ‘atypical-casablancae’ from Essaouira (ES); E, S. glaucus ssp. coronopifolius. All material wild-collected.
Senecio glaucus ssp. coronopifolius seems the most plausible introgressant as this taxon is diploid, has dissected leaves (Fig. 1E) and can occur in maritime dune habitats. In general, S. glaucus ssp. coronopifolius is associated with more xeric habitats in southern Morocco, occurring widely in inland as well as coastal locations. The minimum distance between S. glaucus ssp. coronopifolius and ‘atypical-casablancae’ in the present study was ∼100 km and it remains unclear whether their ranges overlap. Circumstantial support for an introgressive origin is provided by the apparent high fertility of many artificial Senecio hybrids (Alexander 1975). In addition, both putative parental taxa flower concurrently and have unspecialized pollen vectors such as solitary bees (Halictus, Andrena), syrphid flies and other diptera (Abbott & Irwin 1988; Comes & Kadereit 1990). Further, the role of introgression in the origin of other Senecio taxa has been supported by molecular evidence (Abbott et al. 1992a,b, 2000, 2002). Unproven co-occurrence does not by itself rule out hybridization because hybrids can occupy geographical ranges that exclude the parental taxa. For example, where hybrids display higher fitness than one or both of their parents, a broad hybrid zone lacking the parental taxa would be expected (Barton & Hewitt 1985). Alternatively, hybrids may occupy novel habitat (Cruzan & Arnold 1993) or expand into extreme habitat (Lewontin & Birch 1966). In addition, species distributions change over time and currently allopatric species may have been in former contact.
The possibility of restricted gene flow must also be considered as a cause of ‘atypical-casablancae’. Indirect estimates of gene flow in terms of number of migrants (Nm) exchanged among populations using the infinite island model (Wright 1951) have been made in three Mediterranean Senecio species: S. gallicus Vill. from the Iberian Peninsula and S. glaucus and S. vernalis from the Near East (Comes & Abbott 1998, 1999). These species provide a useful comparison with S. leucanthemifolius var. casablancae as all are obligate outcrossers with pappose achenes (fruits) apparently suited to dispersal by wind (Small 1919). Based on nuclear isozyme loci all three display sufficient gene flow to counteract drift and selection (Comes & Abbott 1998, 1999). However, Nm calculated for the haploid chloroplast genome has shown insufficient gene flow to counteract drift and selection in both S. gallicus and S. glaucus (Comes & Abbott 1998, 1999). Further investigation of S. gallicus using an independent nuclear data set composed of randomly amplified polymorphic DNAs (RAPDs) has revealed significant geographical subdivision and isolation by distance (Comes & Abbott 2000). The failure of isozymes to detect this nuclear variation has been ascribed to low mutation rates and/or artefactual uniformity due to small sampling of the genome (Comes & Abbott 2000).
Characteristic patterns of molecular variation provide a means of distinguishing hybridization from divergence. Hybridization would be expected to result in an additive pattern of biparentally inherited markers in at least some individuals. In addition, taxon-specific markers would be expected to show clinal variation across a hybrid zone (Barton & Hewitt 1985). Further, an increase in genetic variation is expected in introgressed populations, due to recombination of divergent nuclear genomes (Arnold 1997) and the generation of mixed populations of nonrecombining organelle genomes (Ennos et al. 1999). In contrast, divergence of populations would be expected to result in unique molecular markers and great differences in marker frequencies.
We have assessed genetic variation in the present study with two DNA-based marker systems: (i) restriction fragment length polymorphism (RFLP) of polymerase chain reaction (PCR)-amplified chloroplast DNA (cpDNA) and (ii) RAPDs (Williams et al. 1990). The chloroplast genome is a valuable source of markers for studying hybridization due to uniparental inheritance and its relatively low variability. Senecio, in common with most angiosperms, shows maternal inheritance of cpDNA (Harris & Ingram 1992). In contrast, RAPDs represent a random sample of the entire genome (Williams et al. 1990). However, due to the small size of the organelle genomes, RAPDs reflect primarily the nuclear genome (Lorenz et al. 1994). The predominant biparental inheritance of RAPDs means that additive patterns of markers are expected in hybrids, and consequently they have been used widely to examine hybridization and introgression (e.g. Sale et al. 1996; Martin & Cruzan 1999).
The aim of the present study was to use cpDNA PCR–RFLP and RAPDs to: (i) test the competing causal hypotheses that ‘atypical-casablancae’ is either the product of introgression between S. glaucus ssp. coronopifolius and S. leucanthemifolius var. casablancae or a product of divergence within S. leucanthemifolius, and (ii) quantify genetic variation across the range of the restricted endemic S. leucanthemifolius var. casablancae including ‘atypical-casablancae’.
Materials and methods
Plant material
Seed was collected from nine wild populations, with S. leucanthemifolius var. casablancae, ‘atypical-casablancae’ and S. glaucus ssp. coronopifolius each represented by three populations (Table 1). Collection localities were, as far as possible, distributed evenly along the Moroccan Atlantic coast, and reflect the entire known range of S. leucanthemifolius var. casablancae and ‘atypical-casablancae’. A single inland population of S. glaucus ssp. coronopifolius was also included. The distance to the nearest populations was on average 81 km (range 31–165 km). In all cases high plant densities enabled each population to be sampled from a radius of less than 1 km. Ten individuals were grown from each population to provide leaf material for DNA extraction. Germination was generally sufficient to allow 10 separate maternal plants to be used from each population. Herbarium specimens for all but two populations were made at the time of seed collection (Table 1).
| Species/population | Pop. code | Latitude | Longitude | Voucher* |
|---|---|---|---|---|
| S. leucanthemifolius var. casablancae | ||||
| Sehb-Eddheb — dune | SE | 33°56′ N | 06°56′ W | Forbes cult. 79–10 |
| El-Jadida — forested dune | JA | 33°15′ N | 08°30′ W | Coleman & Abbott 01/99 |
| Cap Beddouza — sandy clifftop | BE | 32°36′ N | 09°13′ W | Coleman & Abbott 05/99 |
| ‘atypical-casablancae’ | ||||
| Souira Kédima — dune | SK | 32°02′ N | 09°21′ W | Coleman & Abbott 07/99 |
| El-Mehattat — sandy gullies | ME | 31°50′ N | 09°34′ W | Coleman & Abbott 08/99 |
| Essaouira — dune | ES | 31°30′ N | 09°46′ W | Coleman & Abbott 09/99 |
| S. glaucus ssp. coronopifolius | ||||
| Oued Sous — sandy estuary | OS | 30°21′ N | 09°37′ W | Coleman & Abbott 11/99 |
| Tizi Mlil — open sandy woodland | TM | 29°43′ N | 09°00′ W | Forbes cult. 71–1 |
| Sidi Ifni — sandy clifftop | SI | 29°23′ N | 10°11′ W | Coleman & Abbott 13/99 |
- * All herbarium vouchers made from wild material collected by MC and RJA in 1999 except SE and TM, which are cultivated material grown from seed collected by RJA in 1996. Vouchers deposited at the herbarium of the Royal Botanic Garden, Edinburgh (E) except SE and TM, which are at the herbarium of the University of St Andrews (STA).
DNA extraction
Young leaf material was harvested from the greenhouse and total genomic DNA was isolated from ∼100 mg of tissue using a modified hexadecyltrimethylammonium bromide (CTAB) miniprep method (Doyle & Doyle 1990). DNA concentrations were determined, relative to uncut lambda DNA, on 1% agarose gels.
Chloroplast DNA PCR–RFLP
Chloroplast DNA fragment amplifications were carried out in 25 µL total volumes containing 1 unit Biotaq™ polymerase (Bioline), 10% volume 10 × Biotaq buffer (160 mm (NH4)2SO4, 670 mm Tris-HCl, 0.1% Tween-20), 2 mm MgCl2, 0.1 mm dNTPs, 0.2 µm of each of the primers and approximately 5 ng of template DNA. Five primer pairs, HK, CS, SM, AS and ML (Demesure et al. 1995), were used. The thermal cycle was as described by (Demesure et al. 1995) with annealing temperatures and extension times as follows: HK, 62 °C and 2 min; CS, 54 °C and 2 min; SM, 55 °C and 2 min; AS, 55 °C and 4 min; and ML, 55 °C and 4 min amplification products were digested with eight restriction endonucleases: RsaI, PstI, SspI, HaeIII, MspI, BamHI, HinfI and StuI (Promega) according to the manufacturer's instructions. Digested fragments were separated on 8% polyacrylamide gels (acrylamide: bisacrylamide, 37.5:1) and TBE buffer (0.5×). An electric current (constant voltage 300 V, current ∼30 mA per gel) was passed through the gels until the loading dye reached the end of the gels (∼3 h), after which they were stained with ethidium bromide and restriction patterns were visualized by UV transillumination.
RAPD procedure
RAPD reactions were carried out in 25 µL total volumes containing 1 unit Biotaq™ polymerase (Bioline), 10% volume 10 × Biotaq buffer (160 mm (NH4)2SO4, 670 mm Tris-HCl, 0.1% Tween-20), 2.5 mm MgCl2, 0.2 mm dNTPs, 0.2 µm of primer and approximately 5 ng of template DNA. Denaturation at 94 °C for 3 min was followed by a thermal cycle of: 30 s denaturation, 94 °C; 45 s annealing, 35 °C; 1.5 min extension, 72 °C; 44 cycles. A final extension step of 4 min at 72 °C was carried out. PCR products were separated on 1.5% agarose gels containing 0.5 µg/µL ethidium bromide and visualized by UV transillumination. Two hundred primers from the University of British Columbia (RAPD sets nos one and two) were screened using six individuals (two from each of S. leucanthemifolius var. casablancae, ‘atypical-casablancae’ and S. glaucus ssp. coronopifolius). Thirteen primers that gave strong, easily scorable and reproducible bands were selected (UBC-112: GCTTGTGAAC; 141: ATCCTGTTCG; 177: TCAGG CAGTC; 204: TTCGGGCCG; 221: CCCGTCAATA; 226GG GCCTCTAT; 234: TCCACGGACG; 238: CTGTCCAGCA; 240: ATGTTCCAGG; 241: GCCCGACGCG; 248: GAGTA AGCGG; 249GCATCTACCG; 288: CCTCCTTGAC).
Chloroplast DNA analysis
Chloroplast DNA haplotypes were defined by the presence/absence of all detected length and site mutations. All individuals were assigned to a haplotype, and haplotype frequencies were calculated for each population. Differences in haplotype frequency among species were analysed with an R (taxa) × C (haplotypes) test of independence using the G-test (Sokal & Rohlf 1995). Differences in haplotype frequency within S. leucanthemifolius were examined in the same way by comparing var. casablancae with ‘atypical-casablancae’. Nei's (1987) unbiased estimates of haplotypic diversity (h) and their standard errors and the effective number of haplotypes (Ne) were calculated for each population. The significance of among population haplotypic differentiation was assessed by t-test following Nei (1987).
RAPD analysis
A binary data matrix was produced by scoring each RAPD fragment as present or absent for each individual. Interindividual relationships in multidimensional space were examined by principal coordinate (PCO) analysis using PCO3D (Adams 1995). The PCO analysis used a similarity matrix derived from Jaccard's (1908) coefficient: Dj = nxy/n, where nxy is the number of shared markers and n is the total number of markers scored in each pairwise comparison excluding shared absences. PCO analysis is particularly well suited to identifying cases of recent and ongoing hybridization, as hybrids are located generally between their parents in PCO plots (e.g. Sale et al. 1996).
The hierarchical partitioning of genetic variation was estimated using analysis of molecular variance (amova) of squared Euclidean distance between individuals (Excoffier et al. 1992). amova analyses were conducted using arlequin 2.001 (Schneider et al. 2000), thus enabling extraction of variance components and calculation of analogs of F-statistics (Wright 1978) called Φ-statistics. arlequin was also used to calculate pairwise population ΦST as a measure of genetic distance among populations, and to conduct Mantel tests (Mantel 1967) to test if genetic distance (ΦST) among pairs of populations was correlated significantly with geographical distance. Significance levels for all analyses implemented with arlequin were calculated using a nonparametric permutation procedure with 10 000 random permutations.
Shannon's diversity index was chosen to examine further the distribution of diversity within and between species as it does not rely on any assumption of Hardy–Weinberg equilibrium (Chalmers et al. 1992; Yeh et al. 1995). Shannon's diversity index was calculated as: HO =− ΣPi log2Pi, where Pi is the frequency of the ith RAPD band. Diversity was calculated at several hierarchical levels: HO was calculated for each population and then averaged over populations to give HPOP, while HT was calculated as the corresponding value across all populations. Diversity was partitioned into a within-population component as HPOP/HT, and into an among-population component as (HT-HPOP)/HT. This provides both a comparison for results generated by amova and a means of comparing the level of diversity found in individual populations, which is useful from a conservation perspective for identifying areas of particular diversity.
Results
Chloroplast DNA variation
Length variation was revealed most clearly in two digestions: HK digested with HinfI and AS digested with RsaI. In each case a single digestion fragment displayed length variation. The variable HK fragment occurred as four length variants: 225, 230, 245 and 265 base pairs (bp). The variable AS fragment occurred as three length variants: 370, 385 and 395 bp. Combining these length variants for all individuals gave a total of five cpDNA haplotypes, designated A, B, C, D and E (Table 2).
| Species/population | n | Haplotype* | h | SE h | N e | ||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | |||||
| S. leucanthemifolius var. casablancae | |||||||||
| SE | 10 | 0.8 | 0.1 | — | 0.1 | — | 0.358 | 0.127 | 1.515 |
| JA | 10 | 0.8 | 0.2 | — | — | — | 0.337 | 0.110 | 1.471 |
| BE | 10 | 0.9 | — | 0.1 | — | — | 0.189 | 0.108 | 1.220 |
| ‘atypical-casablancae’ | |||||||||
| SK | 10 | 0.9 | — | 0.1 | — | — | 0.189 | 0.108 | 1.220 |
| ME | 10 | 0.5 | — | — | 0.5 | — | 0.526 | 0.036 | 2.000 |
| ES | 10 | 0.6 | 0.1 | — | — | 0.3 | 0.568 | 0.086 | 2.174 |
| S. glaucus ssp. coronopifolius | |||||||||
| OS | 10 | — | — | — | — | 1.0 | 0.000 | 0.000 | 1.000 |
| TM | 10 | — | 0.1 | — | — | 0.9 | 0.189 | 0.108 | 1.220 |
| SI | 10 | — | — | — | — | 1.0 | 0.000 | 0.000 | 1.000 |
| Total | 90 | 0.500 | 0.055 | 0.022 | 0.066 | 0.355 | |||
- * Haplotypes defined by the following fragment sizes: A, HK 245-bp AS 385-bp; B, HK 245-bp AS 395-bp; C, HK 230-bp AS 395-bp; D, HK 265-bp AS 370-bp; E, HK 225-bp AS 395-bp.
A marked division in haplotype frequency according to species was seen. The E haplotype was fixed in the two coastal populations of S. glaucus ssp. coronopifolius and nearly so (0.9) in the single inland population. In contrast, this haplotype was absent from five of six populations of S. leucanthemifolius (Fig. 2, Table 2). The single occurrence of haplotype E in S. leucanthemifolius was in the population (ES) geographically closest to S. glaucus ssp. coronopifolius, suggesting that this may result from introgression. Similarly, haplotype A was found to occur at high frequency in both S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ but was absent from S. glaucus ssp. coronopifolius. This apparent difference in haplotype frequencies among species was highly significant (G = 89.66, d.f. = 4, P < 0.001, G-test). Haplotypic structuring was not significant between S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ (G = 8.67, d.f. = 4, P > 0.05, G-test).
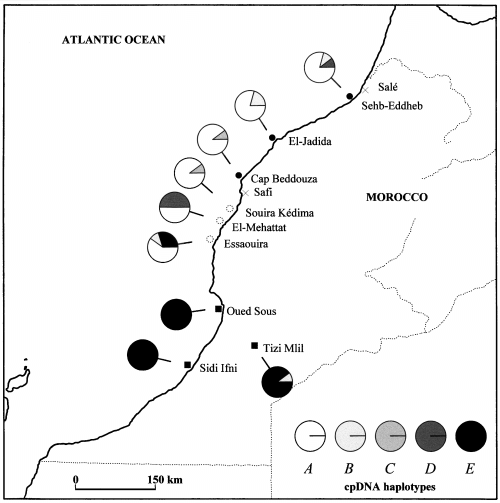
Map of frequencies of five chloroplast DNA (cpDNA) haplotypes observed in Senecio leucanthemifolius var. casablancae (filled circles), ‘atypical-casablancae’ (open circles) and S. glaucus ssp. coronopifolius (filled squares).
Haplotypic polymorphism was observed in all six populations of S. leucanthemifolius and one of three populations of S. glaucus ssp. coronopifolius. The effective number of haplotypes (Ne) within polymorphic populations ranged from 1.471 to 2.174, and haplotypic diversity ranged from 0.189 to 0.568 (Table 2). Haplotypic diversity was greatest in the two most southerly populations (ME, ES) of ‘atypical-casablancae’, followed by the most northerly population (SE) of S. leucanthemifolius var. casablancae. Pairwise comparisons of haplotypic diversity among populations of S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ revealed that the two most diverse populations (ME, ES) were significantly different from the two least diverse populations (BE, SK) (t = 2.74 and 2.96, P < 0.01, t-test). Otherwise differences in diversity were nonsignificant.
RAPD variation
Twenty-eight polymorphic RAPDs were amplified using 13 primers and scored in 90 individuals. The number of fragments scored per primer ranged from one (UBC-141, 221) to three (UBC-234, 238, 241, 288). All 90 individuals possessed unique RAPD phenotypes. Only a single diagnostic marker (defined as occurring in all individuals of taxon A and none of taxon B) was identified. This marker (UBC-288/1) was fixed in both S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ and absent in S. glaucus ssp. coronopifolius. Taxon specific markers (defined as occurring in some individuals of taxon A and none of taxon B) were more numerous. Three markers (UBC-112/2, 234/2, 240/2) were specific to S. glaucus ssp. coronopifolius, although all occurred at low frequency (= 0.3). All three were also absent from ‘atypical-casablancae’. Three markers (UBC-141/1, 234/3, 288/2) were specific to S. leucanthemifolius var. casablancae, ranging in frequency from 0.3 to 0.6. All three occurred in ‘atypical-casablancae’ at almost identical frequency to that observed in S. leucanthemifolius var. casablancae. Two markers (UBC-226/1, 241/1) displayed high frequency (defined as > 0.75) in S. glaucus ssp. coronopifolius and low frequency (defined as < 0.25) in S. leucanthemifolius var. casablancae. Both occurred in ‘atypical-casablancae’ at identical, or nearly so, frequency to that observed in S. leucanthemifolius var. casablancae.
PCO analysis was used to examine the relationships among individuals in multidimensional space. The first three axes of the PCO analysis accounted for 21.23, 8.09 and 5.16% of the total variance, respectively. Plotting PCO1 against PCO2 clearly distinguished two discreet clusters corresponding to S. leucanthemifolius and S. glaucus ssp. coronopifolius (Fig. 3), indicating that these species represent distinct genetic entities. A division was also evident between S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ (Fig. 3). There was only slight overlap between these subclusters, clearly indicating the presence of genetic differentiation. Plotting PCO1 against PCO3 revealed no additional relationships (not shown).
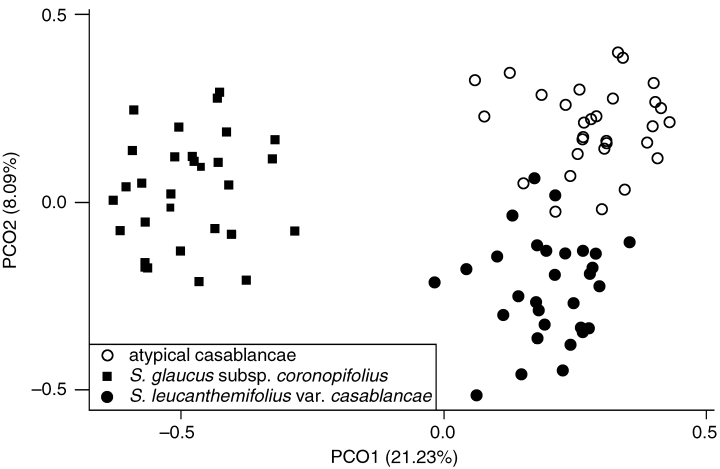
Principal coordinates (PCO) analysis plot of the first two axes calculated using Jaccard's (1908) distance based on 28 random amplified polymorphic DNA (RAPD) fragments in Senecio leucanthemifolius var. casablancae, ‘atypical-casablancae’ and S. glaucus ssp. coronopifolius.
Genetic differentiation between species and within S. leucanthemifolius was also revealed by estimation of variance components and Φ-statistics with amova (Table 3). Three analyses were conducted in which alternative hierarchical groupings were used to evaluate genetic structure (Table 3). Based on the three clusters identified by PCO the variance component was large and significant within populations (52.67%, P < 0.001; Table 3) and among clusters (39.70%, P = 0.004; Table 3). Among populations the variance component was relatively small, but significant (7.63%, P < 0.001; Table 3). Defining the top level of the hierarchy as the two species produced an almost identical result (Table 3). However, defining S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ as the top level of the hierarchy produced an increase in the variance component within populations (69.94%, P < 0.001; Table 3) but little change among populations (10.90%, P < 0.001; Table 3). In addition, the variance component at the top level of the hierarchy was reduced by about half and not significant (19.16%, P = 0.10; Table 3). Individuals of both species were differentiated significantly from each other (ΦCT = 0.41, P = 0.01; Table 3), while differentiation of S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ was significant only at the 10% level (ΦCT = 0.19, P = 0.10; Table 3).
| Source of variation | d.f. | SSD | Variance component | % of total | Fixation indices | P-value |
|---|---|---|---|---|---|---|
| Analysis 1 | ||||||
| Among clusters | 2 | 163.91 | 2.46 | 39.70 | ΦCT = 0.40 | = 0.004 |
| Among populations | 6 | 48.07 | 0.47 | 7.63 | ΦSC = 0.13 | < 0.001 |
| Within populations | 81 | 264.90 | 3.27 | 52.67 | ΦST = 0.47 | < 0.001 |
| Analysis 2 | ||||||
| Among species | 1 | 127.69 | 2.89 | 41.08 | ΦCT = 0.41 | = 0.01 |
| Among populations | 7 | 84.23 | 0.88 | 12.46 | ΦSC = 0.21 | < 0.001 |
| Within populations | 81 | 264.90 | 3.27 | 46.46 | ΦST = 0.54 | < 0.001 |
| Analysis 3 | ||||||
| Among groups | 1 | 36.22 | 0.92 | 19.16 | ΦCT = 0.19 | = 0.10 |
| Among populations | 4 | 34.40 | 0.52 | 10.90 | ΦSC = 0.13 | < 0.001 |
| Within populations | 54 | 181.50 | 3.36 | 69.94 | ΦST = 0.30 | < 0.001 |
- Degrees of freedom (d.f.), sum of squared deviations (SSD) and the significance (P) of the variance components and fixation indices are shown. Levels of significance are based on 10 000 permutations.
Thirty-four of the 36 pairwise genetic distances (ΦST) between populations were significant (Table 4). In both cases where the genetic distance was not significant the geographical distances between populations were relatively low [71 km between Souira Kédima (SK) and Essaouira (ES), and 93 km between Oued Sous (OS) and Tizi Mlil (TM)]. In general, genetic distances and significance levels were lowest for pairwise comparisons within each of the three groups (Table 4).
| Pop. | SE | JA | BE | SK | ME | ES | OS | TM |
|---|---|---|---|---|---|---|---|---|
| SE | — | |||||||
| JA | 0.27*** | — | ||||||
| BE | 0.17*** | 0.11** | — | |||||
| SK | 0.35*** | 0.31*** | 0.26*** | — | ||||
| ME | 0.35*** | 0.31*** | 0.30*** | 0.10* | — | |||
| ES | 0.33*** | 0.26*** | 0.20*** | 0.06 | 0.07* | — | ||
| OS | 0.48*** | 0.54*** | 0.46*** | 0.55*** | 0.57*** | 0.56*** | — | |
| TM | 0.49*** | 0.53*** | 0.44*** | 0.51*** | 0.52*** | 0.52*** | 0.04 | — |
| SI | 0.53*** | 0.59*** | 0.54*** | 0.56*** | 0.57*** | 0.59*** | 0.18** | 0.11** |
- * P < 0.05;
- ** P < 0.01;
- *** P < 0.001. P-values indicate the probability that a random genetic distance (ΦST) is larger than the observed distance and are based on 10 000 permutations.
Testing the hypothesis of isolation by distance for the complete data set showed that the correlation between genetic distance (ΦST) and geographical distance between pairs of populations was positive and highly significant (r = 0.64, P < 0.001, Mantel-test). However, the clear differentiation of S. leucanthemifolius and S. glaucus ssp. coronopifolius may have biased this result. Nevertheless, in the data set consisting of S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ the correlation between genetic and geographical distances was also positive and significant (r = 0.68, P = 0.01, Mantel-test).
Shannon's diversity index calculated for S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ ranged from 0.32 for Essaouira to 0.37 for Cap Beddouza and Souira Kédima, with a mean value (HPOP) of 0.35 (Table 5). No significant differences in diversity level were found between populations. The within-population diversity component was 70%, whereas 30% was maintained among populations (Table 5). These figures are identical to those derived from amova (Table 3 analysis 3).
| Pop. | H O | H POP | H T | H POP/HT | H T-HPOP/HT |
|---|---|---|---|---|---|
| BE | 0.37 | ||||
| SK | 0.37 | ||||
| SE | 0.36 | ||||
| JA | 0.35 | ||||
| ME | 0.35 | ||||
| ES | 0.32 | ||||
| 0.35 | 0.50 | 0.70 | 0.30 |
Discussion
Hybridization and introgression
No evidence was obtained from RAPD analysis that hybridization occurs between S. leucanthemifolius var. casablancae and S. glaucus ssp. coronopifolius along the Moroccan Atlantic coast. Additivity of RAPD taxon markers was not observed and diversity levels were uniform across the range of var. casablancae and the morphologically divergent ‘atypical-casablancae’. Moreover, PCO analysis neither revealed a genetic continuum between the two species, nor located ‘atypical-casablancae’ between the putative parents as might be expected if it were of hybrid origin. The distinct genetic identities of S. leucanthemifolius var. casablancae sensu lato and S. glaucus ssp. coronopifolius were also highlighted by the significant differentiation between species found by amova (ΦCT = 0.41, P = 0.01; Table 3). The RAPD analysis suggests therefore that either there are strong barriers to reproduction or that these taxa are allopatric. The latter situation is possible as the minimum distance observed between ‘atypical-casablancae’ and S. glaucus ssp. coronopifolius in the present study was ∼100 km.
A possible concern with interspecific studies using RAPDs is that comigrating fragments from different species are not always homologous (Rieseberg 1996). In the present study this problem should be minimized by the close phylogenetic relationship between S. leucanthemifolius and S. glaucus (Comes & Abbott 2001). However, false homologies would only reduce the genetic distance between taxa, thereby strengthening our conclusion that S. leucanthemifolius var. casablancae sensu lato and S. glaucus ssp. coronopifolius represent distinct genetic entities.
The cpDNA analysis (Fig. 2, Table 2) largely supports the genetic isolation of the species. However, limited sharing of haplotypes B and E was observed. This could be interpreted as evidence of past hybridization. However, shared haplotypes may also arise by convergence. Further, as many plant species are thought to be paraphyletic shortly after speciation (Rieseberg & Brouillet 1994), shared haplotypes may reflect incomplete lineage sorting of ancestral polymorphism.
The E haplotype occurred in ‘atypical-casablancae’ at Essaouira (ES) at moderate frequency (0.3) but was otherwise restricted to S. glaucus ssp. coronopifolius. Convergence seems an unlikely explanation, because this would require two independent mutations. Incomplete lineage sorting could explain haplotype sharing in this case, particularly as divergence time estimates for these taxa are relatively low at 0.44–0.88 million years ago (Comes & Abbott 2001). However, we favour hybridization as an explanation based upon the geographical distribution of the E haplotype. In a situation of incomplete lineage sorting the E haplotype in S. leucanthemifolius would not be expected to show any geographical relationship with S. glaucus ssp. coronopifolius. The clear clustering of the E haplotype (Fig. 2) means that hybridization provides the simplest explanation. In contrast, the B haplotype shows no obvious clustering. This haplotype occurs at low frequency (= 0.2) in two populations (SE, JA) of S. leucanthemifolius var. casablancae, one population (ES) of ‘atypical-casablancae’ and the only inland population (TM) of S. glaucus ssp. coronopifolius. Consequently, incomplete lineage sorting may best explain haplotype sharing in this case.
If hybridization is the cause of the observed sharing of the E haplotype, S. glaucus ssp. coronopifolius would have acted as maternal parent in past hybridization events, given the maternal inheritance of cpDNA in Senecio (Harris & Ingram 1992). The apparent absence of nuclear markers of S. glaucus ssp. coronopifolius in the Essaouira (ES) population indicates that backcrossing to S. leucanthemifolius would have occurred repeatedly and produced a nuclear genome largely characteristic of the latter. Such ‘chloroplast capture’ has been found to be widespread in plants (Rieseberg & Soltis 1991).
The absence of detailed distribution data means that the circumstances under which chloroplast capture could have occurred remain speculative. If these taxa are parapatric, with little range overlap, the most probable cause of this pattern is rare establishment of S. glaucus ssp. coronopifolius in S. leucanthemifolius populations, possibly via long-distance achene dispersal. In this situation, it is probable that genetic swamping by the numerically larger S. leucanthemifolius would lead to the elimination of the S. glaucus nuclear genome. The alternative of high levels of long-distance pollen flow from S. leucanthemifolius seems unlikely, as a study of pollinator foraging behaviour in North American Senecio has indicated that pollen will not easily move more than 50–100 m (Schmitt 1980). Alternatively, if range overlap is large and mixed populations exist (or have existed), hybrid swarms containing a mixture of the two nuclear genomes may occur. Although no evidence for hybrid swarms has been found, this possibility cannot be ruled out as many areas of coastline proved to be largely inaccessible. It is possible that a S. glaucus haplotype could be captured in the manner outlined above by long-distance dispersal from a hybrid zone.
An important conclusion from these results is that, although past hybridization may have occurred, the RAPD data indicate introgression from S. glaucus ssp. coronopifolius is not a potential threat to the genetic integrity of S. leucanthemifolius var. casablancae.
Genetic structure ofSenecio leucanthemifoliusvar.casablancae
Various life-history traits in plants such as the vagility of pollen and seeds and breeding system are linked strongly to the development of genetic structure in allozymes (Hamrick & Godt 1989; Hamrick & Godt 1996) and RAPDs (Nybom & Bartish 2000). Generally, long-lived, outcrossing, late successional taxa retain the majority of genetic variation within populations, while annual, selfing, early successional taxa allocate the majority of variation among populations. As an obligate outcrosser with apparently high potential for seed dispersal S. leucanthemifolius may be expected to have a relatively homogeneous distribution of variation among populations, with the majority of variation maintained within populations. The present study supports this in as far as the majority of variation is maintained within populations (69.94%, P < 0.001; Table 3). However, genetic variation does not show a homogeneous distribution and population differentiation is evident. A small but significant amount of variation is maintained among populations (10.90%, P < 0.001; Table 3), while variation maintained among S. leucanthemifolius var. casablancae and ‘atypical-casablancae’ is significant at the 10% level (Table 3). Genetic distances as assessed by pairwise ΦST were significant in all but one of the 15 comparisons in S. leucanthemifolius (Table 4). In addition, the significant positive correlation of genetic and geographical distances (r = 0.68, P = 0.01) indicates that gene flow is restricted, with populations fitting the isolation by distance model. The overall value of ΦST was 0.30, which suggests a high level of population structure. This value is close to the average value of 0.28 reported for outcrossing taxa by Nybom & Bartish (2000) in their review of amova-derived ΦST. It is also close to the value of 0.34 recorded in the related outcrosser S. gallicus (Comes & Abbott 2000).
These results suggest that restricted gene flow has resulted in significant population differentiation. Small-scale genetic differentiation has been documented widely in plants (see Linhart & Grant 1996; for a review). However, taxa with seeds ingested by animals or dispersed by wind or water have been found to display significantly lower population differentiation than taxa with gravity or adhesive dispersal (Nybom & Bartish 2000). Because the pappose achenes in S. leucanthemifolius are assumed to aid wind dispersal, the highly localized nature of population differentiation is unexpected. Indeed, experimental work on the closely related S. vulgaris L. has indicated that under favourable conditions there is effectively no limit to seed dispersal (Small 1919). Our data contradict this view, indicating instead that long-distance seed dispersal is rare in S. leucanthemifolius.
Restricted gene flow is consistent with the remarkable level of morphological differentiation displayed by the three populations of ‘atypical-casablancae’ (Fig. 1). The apparent rarity of long-distance seed dispersal is also understandable on a more critical examination of the likely dispersal benefits of the pappus. The achenes of both species in this study shed their pappus readily at maturity. This characteristic is common to most members of sect. Senecio from the Mediterranean that we have examined. Consequently, the pappus may not be a particularly effective means of long-distance dispersal. This finding may be relevant to understanding the complicated pattern of morphological variation in S. leucanthemifolius as a whole. Restricted gene flow may also be an important factor in the evolution of other members of the Mediterranean species complex. It is notable that a similar, but morphologically less marked, pattern of intraspecific differentiation has been found in S. gallicus in the Iberian Peninsula and southern France (Comes & Abbott 2000). Nevertheless, isolation by distance in the present study indicates that some long-distance dispersal has taken place, as without it a significant correlation between genetic and geographical distance would not exist.
The coast between Cap Beddouza and Safi is largely rocky and lacking suitable dune habitats. This apparently unsuitable habitat separates var. casablancae from ‘atypical-casablancae’. In light of the seemingly limited dispersal ability of achenes this may present a natural barrier to gene flow that has favoured divergence.
It should be stressed that environmental variation between sites may be partly responsible for the morphological variation observed in the field. There is a marked rainfall gradient running along the coast and exposure varies due to topography and shrub cover. The leaf morphology differences between populations of ‘atypical-casablancae’ appear to be reduced under cultivation, with the lobing of leaves becoming more pronounced. However, the unlobed condition of S. leucanthemifolius var. casablancae is not lost under cultivation, indicating that this characteristic has a large component of genetic control.
Taxonomic implications and future work
The morphological and genetic distinction of ‘atypical-casablancae’ from S. leucanthemifolius var. casablancae means that consideration should be given to the value of formally recognizing this variation. In the most recent revision of the Mediterranean members of sect. SenecioAlexander (1979) noted that for S. leucanthemifolius. ‘It is difficult to make satisfactory infraspecific groups in this extremely variable species’. A particular problem is that some varieties appear to lack clear boundaries due to continuous variation interconnecting them (Alexander 1979). This is not the case with ‘atypical-casablancae’, which instead simply represents undescribed variation within a distinct entity, S. leucanthemifolius var. casablancae.
Briefly, considering the species-wide problem of complex and intergrading variation, one solution would be to recognize S. leucanthemifolius as a polymorphic species with no infraspecific taxa. We do not advocate this approach because it would obscure valuable biological information. For instance, an important division exists between erect forms of S. leucanthemifolius in the west Mediterranean and procumbent fleshy-leaved forms, which include var. leucanthemifolius, in the central and east Mediterranean.
Based purely on morphology, S. leucanthemifolius var. casablancae may be closely allied to the Canary Island endemic S. bollei Sunding & Kunkel. Both exhibit an erect habit of growth and more or less simple rhomboid leaves. The relationship between these taxa should be investigated as a closer link to S. bollei than S. leucanthemifolius sensu stricto may indicate that species status is more appropriate for var. casablancae. Regardless of the relationship to S. bollei, the morphological and ecological distinctness of S. leucanthemifolius var. casablancae from other varieties of S. leucanthemifolius by itself suggests that species rank may be appropriate. We advocate species status but acknowledge that it is a somewhat subjective distinction between specific and infraspecific rank. An advantage of treatment at specific rank is that the variation encompassed by ‘atypical-casablancae’ could be recognized at infraspecific level.
Taxonomic changes should await a revision of S. leucanthemifolius and closely allied species. This is necessary because our field observations and examination of herbarium material indicate that in addition to the variation examined in the present study Morocco supports further variation that is not encompassed by Alexander's (1979) varietal classification. Most importantly, none of the Moroccan material we have seen corresponds to the procumbent fleshy-leaved form of S. leucanthemifolius sensu stricto. This view is contrary to Alexander (1979), who recorded var. leucanthemifolius along the North African coast from Tunisia to Morocco and down the Moroccan Atlantic coast. The apparent geographical separation of the two growth forms suggests that they may represent genetically distinct groups. Consequently, the taxonomic rank of the other two erect growing west Mediterranean varieties, var. major Ball and var. fradinii (Pomel) Batt., should also be reassessed. These varieties would also benefit from revision as they intergrade with each other.
In conclusion, further morphological and molecular work is required to clarify relationships within the S. leucanthemifolius complex as a whole. Most importantly, the significance of the division of erect and procumbent growth forms between west and central/east Mediterranean populations, respectively, needs to be assessed. These different growth forms are particularly marked in cultivated material. By focusing on variation in a morphologically discreet variety of S. leucanthemifolius the present study has highlighted the importance of restricted gene flow in promoting localized divergence. It seems likely that localized divergence is also a factor responsible for the wider taxonomic problems of S. leucanthemifolius. Finally, an interesting comparison may be made with related taxa farther north. Senecio rupestris Waldst. & Kit., for instance, occupies a much larger range than S. leucanthemifolius var. casablancae and yet shows comparatively little variation in morphology (Abbott et al. 2002). This difference may reflect the relatively recent colonization of more northern latitudes after the last glacial period ended ∼10 000 years ago. In light of this, it would be interesting to establish the timeframe of divergence both in var. casablancae and in S. leucanthemifolius as a whole.
Acknowledgements
We thank David Forbes for assistance in the cultivation of plant material, Amanda Gillies for invaluable technical advice and Natalie Hueber for conducting a preliminary study. This research was supported by a BBSRC research studentship to MC.
References
This research forms part of a PhD undertaken by Max Coleman. The objective was to use molecular tools to evaluate the systematics, phylogeography and evolution of the taxonomically difficult Senecio sect. Senecio. Work has focused primarily on the Mediterranean Basin, which is a centre of diversity for this group. Richard Abbott's research programme at the University of St Andrews focuses on general issues of plant population genetic structure, processes of speciation, genome composition and the reconstruction of plant phylogenies.




