The recent evolutionary origin of the phenotypically novel amphipod Hyalella montezuma offers an ecological explanation for morphological stasis in a closely allied species complex
Abstract
Numerous molecular studies have identified morphologically cryptic, freshwater invertebrate species, but have not suggested possible mechanisms for their phenotypic stasis. The amphipod crustacean genus Hyalella contains numerous morphologically cryptic species in the H. azteca complex, as well as a small number of morphologically very divergent, narrowly endemic taxa. One such taxon, Hyalella montezuma, is the sole planktonic filter-feeder within the North American amphipod fauna, and is known only from Montezuma Well, a fishless travertine spring mound in Arizona, USA. In this study, we conduct a phylogenetic analysis of mtDNA sequence data using likelihood, Bayesian and cladistic approaches to determine both the relationship of H. montezuma to the H. ‘azteca’ species complex, and to ascertain if its morphological and ecological differentiation have been comparatively recent. The results show that H. montezuma has a very close phylogenetic affiliation with one lineage in the H. azteca complex, indicating that its origin has been recent. We present evidence suggesting that fish predation is an important ecological factor, which constrains morphological and ecological diversification within the genus Hyalella, and that Montezuma Well has provided a relaxation on this constraint.
Introduction
Until recently, a temporal context for the diversification of most extant organisms could only be gained through comparative morphological studies across geographical barriers of known age. However, molecular studies are now providing new and unexpected insights into the temporal pattern of cladogenesis and speciation events. Among freshwater invertebrates, these studies have identified striking discordances between levels of morphological and molecular diversification. Morphologically cryptic, but highly genetically divergent species, are common (Väinöla et al. 1994; King & Hanner 1998; Taylor et al. 1998; Govedich et al. 1999; Lee 2000; Müller 2000; Witt & Hebert 2000; Gómez et al. 2002), providing evidence that many speciation events are ancient, and were accompanied by little or no morphological change. However, few investigators have attempted to ascertain the factors that maintain morphological stasis among cryptic taxa. One mechanism frequently invoked to explain such stasis is stabilizing or normalizing selection, where sister species are subject to similar selective regimes across their ranges. A relaxation in such selective regimes could allow ecological and morphological release, enabling rapid phenotypic change. There are now numerous examples of how changes in the selective environment have provoked phenotypic diversification among morphologically differentiated taxa (Schluter 2000), but how can the selective factors which constrain phenotypic diversification among morphologically cryptic taxa be identified? This is a much more difficult problem because in the case of morphologically cryptic taxa, selective environmental (extrinsic) constraints cannot easily be distinguished from developmental (intrinsic) constraints (Kauffman 1993; Schlichting & Pigliucci 1998). However, comparative studies along habitat gradients can be useful in identifying potential extrinsic agents which maintain morphological similarities among cryptic taxa.
Freshwater habitats vary along two major axes distinguished by permanence and flow regimes, with the extremes being large permanent lakes and small temporary streams. Taxa which occur across this continuum are ideal for studies that aim to determine the effect of environmental variables on the stabilization or diversification of phenotypes. However, very few species and genera are distributed in this way, especially with respect to permanence because of large fitness trade-offs between traits associated with organisms inhabiting temporary and permanent habitats (Wellborn et al. 1996). The amphipod crustacean genus Hyalella (Hyalellidae, Talitroidea) is a useful focus for comparative studies because it includes both morphologically cryptic and morphologically/ecologically highly differentiated species. Hyalella azteca is probably North America's most widely distributed freshwater benthic invertebrate, residing in lotic and lentic permanent habitats stretching from the Atlantic to the Pacific and from Central America to the Beaufort Sea. Morphological investigations on this taxon over a broad geographical range have recognized only a single species (Bousfield 1958, 1996). However, comparative ecological studies along a habitat gradient revealed a size polymorphism between pond and lake dwelling populations, and suggested that predation regimes are responsible for its maintenance (Wellborn 1994, 1995; McPeek & Wellborn 1998). In addition, studies employing allozymes and randomly amplified polymorphic DNA (RAPD) markers revealed considerable genetic divergence among H. ‘azteca’ populations, suggesting that it is a species complex (Thomas et al. 1997, 1998; Hogg et al. 1998; McPeek & Wellborn 1998; Duan et al. 2000). A recent study of allozyme and mtDNA sequence variation among H. ‘azteca’ populations in central glaciated North America revealed a complex of seven reproductively isolated, deeply divergent species (Witt & Hebert 2000). All but one of these seven species are morphologically cryptic, and they exhibit habitat partitioning among ponds and lakes in a way which is consistent with the results of ecological work (e.g. Wellborn 1995).
H. montezuma is endemic to a single habitat- Montezuma Well, Arizona, USA, where it coexists with a member of the H. azteca species complex. H. montezuma is morphologically very divergent from other North American hyalellid amphipods, and is particularly intriguing because it is the sole planktonic, filter-feeder among the more than 210 amphipod species representing 23 genera in North America fresh waters (Cole & Watkins 1977; Blinn & Johnson 1982; Wagner & Blinn 1987). Montezuma Well, a collapsed travertine spring mound, is itself peculiar among North American aquatic habitats. Water entering the well through fissures at its bottom is abnormally rich in carbonates, which dissolute upon entering the well resulting in abnormally high carbon dioxide concentrations ranging from 550 to 600 mg/L (e.g. O’Brien & Blinn 1999).
Prior genetic studies have shown that H. montezuma and the coexisting member of the H. azteca complex exhibit considerable divergence at RAPD loci, suggesting that they were reproductively isolated prior to their colonization of Montezuma Well (Thomas et al. 1994, 1997). In addition, an allozyme study suggested that H. montezuma has a closer genetic affinity with an H. ‘azteca’ population in Ohio than several H. ‘azteca’ populations have with each other (Duan et al. 2000). In this study, we conduct a phylogenetic analysis on mtDNA sequences derived from individuals of H. ‘azteca’ in Arizona and Nevada USA, and from the seven Hyalella species from glaciated North America considered earlier by Witt & Hebert (2000), to establish both the relationship of H. montezuma to the H. azteca species complex, and to provide a temporal context for its evolution. Specifically, we test the hypothesis that the ecological and morphological differentiation of H. montezuma has been recent. This hypothesis would be corroborated by a phylogenetically close association of H. montezuma to any lineage within the H. azteca species complex. Support for the null hypothesis, that H. montezuma is a divergent monophyletic lineage, clearly outside all of the known H. ‘azteca’ lineages, would not permit two possibilities to be distinguished. In this case, H. montezuma might represent a species whose morphological and ecological divergence was ancient, or a lineage whose morphological and ecological differentiation have been recent, but that the lineage with which it shares a most recent common ancestry was not represented in this study.
Materials and methods
Collections
Hyalella montezuma and H. ‘azteca’ were collected in Montezuma Well, Arizona, USA, in March 2001. H. ‘azteca’ was also collected from six additional habitats in Arizona, and a single habitat in Nevada during July 2001 (Table 1). All individuals were preserved and stored in 95% ethanol.
| Location | n | Latitude (N) | Longitude (W) |
|---|---|---|---|
| Montezuma Well AZ | H. montezuma 8 (3) H. ‘azteca’ 12 (5) | 34°39′ | 111°45′ |
| Black River AZ | 8 (7) | 33°51.1′ | 109°18.9′ |
| Bubbling Springs AZ | 5 (2) | 34°46.4′ | 111°54.1′ |
| Crescent Lake AZ | 6 (2) | 33°55.0′ | 109°25.4′ |
| Marshall Lake AZ | 6 (1) | 35°00.0′ | 111°00.3′ |
| Rainbow Lake AZ | 6 (2) | 34°09.5′ | 109°58.9′ |
| Unnamed Pond AZ | 6 (6) | 34°04.0′ | 109°33.1′ |
| Comins Lake NV | 5 (1) | 39°09.4′ | 114°48.8′ |
DNA sequence analysis
Specimens were soaked in ultra pure water at 4 °C for 12–24 h immediately prior to DNA extraction. Total DNA was extracted from each of 5–12 individuals per population by grinding a periopod in 50 µL of a proteinase K extraction buffer (Schwenk 1996). DNA from a specimen of the amphipod Orchestia uhleri (Talitroidea, Talitridae), collected near St. Andrews New Brunswick, Canada, was also extracted for use as an outgroup. A 680 base pair fragment of the mitochondrial cytochrome c oxidase I (COI) gene was amplified using the primers LCO1490 and HCO2198 described by Folmer et al. (1994). The 50 µL PCR reactions contained 0.5 µL (out of 50 µL) of DNA template, 5.0 µL 10× PCR buffer (MBI Fermentas), 0.2 mm of each primer, 2.2 mm MgCl2, 0.2 mm of each dNTP, and 1 unit of Taq DNA polymerase. The PCR conditions consisted of 1 min at 94 °C followed by 5 cycles of 60 s at 94 °C, 90 s at 45 °C, 60 s at 72 °C; followed by 35 cycles of 60 s at 94 °C, 90 s at 51 °C, 60 s at 72 °C; followed by 5 min at 72 °C. PCR products were gel purified (2% agarose) using the Qiaex kit (Qiagen Inc.) and sequenced using the ABI prism BigDye terminator 3 sequencing kit (30 cycles, annealing at 55 °C). Products were sequenced in one direction using primer LCO1490. Electrophoresis was conducted on an ABI 377 automated sequencer (Applied Biosystems). The sequences were aligned by eye using the SeqApp 1.9 sequence editor (Gilbert 1992). Amino acid sequence translations (invertebrate mitochondrial code), mean pairwise transition/transversion ratios, and nucleotide (Tamura & Nei 1993 model) and amino acid (p-distance) distance matrices for all unique sequences obtained in this study, as well as the 27 Hyalella haplotypes representing the seven species identified by Witt & Hebert (2000), were conducted in MEGA 2.1 (Kumar et al. 2001).
Phylogenetic analyses
A preliminary phenetic analysis was conducted using the neighbour-joining (NJ) method and the Tamura & Nei (1993) nucleotide substitution model in MEGA 2.1 to select a reduced set of haplotypes for inclusion in the phylogenetic analyses. As a consequence of the computational requirements for maximum likelihood methods, a subset of haplotypes was selected to represent the haplotype groupings within the major clusters identified by NJ in all subsequent phylogenetic analyses. The program modeltest version 3.0 (Posada & Crandall 1998) was employed to determine the best fit model of nucleotide substitution among the ingroup sequences and O. uhleri (Huelsenbeck & Crandall 1997). modeltest performs a series of hierarchical likelihood ratio tests (Bonferroni corrected) to determine the best fit model of substitution among 56 candidates. The ML analysis was subsequently conducted in paup version 4.0b10 (Swofford 2001), using the model and parameters estimated by modeltest. The ML analysis employed a heuristic search consisting of 20 replicates with taxa added randomly, and tree bisection-re-connection branch swapping with the Multrees and steepest descent options invoked. Confidence in the likelihood analysis was assessed by nonparametric bootstrapping. Again, because of the computational requirements for ML, the bootstrap analysis was conducted with 500 pseudoreplicates, consisting of a single heuristic search per replicate.
A Bayesian phylogenetic analysis (e.g. Huelsenbeck et al. 2001; Lewis 2001) was conducted with mrbayes 2.0 (Huelsenbeck & Ronquist 2001) using the model of nucleotide substitution and parameters estimated by modeltest. Each of four Markov chains were started from random trees and run for one million generations, with the first 15 000 generations (150 trees) discarded as the burn-in. The analysis was run independently 3 times and monitored to ensure likelihood stationarity had been achieved within 15 000 generations.
A maximum parsimony (MP) analysis was conducted in paup with a heuristic search consisting of 10 000 replicates with taxa added randomly, and tree bisection re-connection branch swapping with the steepest decent and MulTrees options invoked. Confidence in the cladistic analysis was assessed a priori by estimating the g1 skewness statistic (Hillis & Huelsenbeck 1992), and a posteriori with a bootstrap analysis employing 2000 pseudoreplicates, with each pseudoreplicate consisting of 2 full heuristic searches and random addition of taxa.
Results
Sixty-two COI sequences were obtained for Hyalella individuals from populations in Arizona and Nevada (Table 1). The 637 base pair sequence alignment and amino acid sequence translations were unambiguous, as there were no gaps or nonsense codons among the 29 unique haplotypes identified in this study. All of these sequences, as well as the 27 Hyalella haplotypes representing the seven species identified by Witt & Hebert (2000), are available in GenBank (accession numbers AY152751-AY152807). Among the 57 sequences (56 Hyalella haplotypes and the outgroup), 278 nucleotide sites were variable.
The preliminary NJ analysis indicated that each of the 29 haplotypes from Arizona and Nevada was associated with one of the seven species (clades) identified by Witt & Hebert (2000) (Fig. 1). The H. ‘azteca’ populations in the Unnamed pond and Marshall Lake had a close phenetic association with clade 1, while the H. ‘azteca’ population in Comins Lake NV clustered with clade 2. H. montezuma also clustered with clade 2, showing an average of 5.4 ± 0.8% sequence divergence from other members of this group. Two lineages were detected in Rainbow Lake which clustered with clades 6 (5 individuals) and 1(a single individual). The H. ‘azteca’ populations in Montezuma Well, Crescent Lake, and the Black River clustered with clade 7. Individuals from Bubbling Springs also had a phenetic affiliation with clade 7, but were considerably divergent, exhibiting a minimum of 15% sequence divergence from any other haplotype within this group. A single haplotype in the Black River (BlaA) showed a minimum of 9.1% sequence divergence from all other haplotypes detected in this population. Average pairwise nucleotide sequence divergence between haplotypes in different clusters ranged from a low of 8.5% to a maximum of 27.5%, while the amino acid sequence divergence among clusters was as high as 7.9% (Table 2). This data set was reduced to a set of 26 ingroup members and the outgroup for all subsequent analysis.
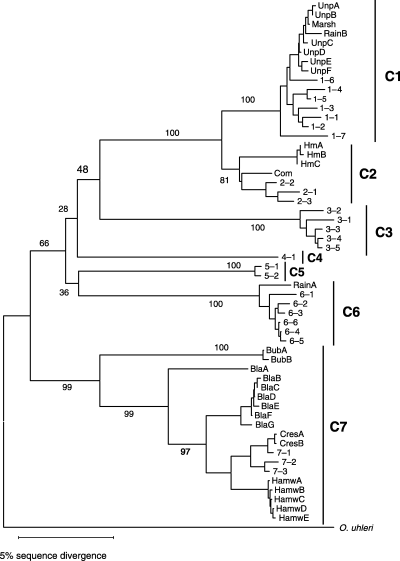
NJ phenogram showing phenetic relationships among 57 COI haplotypes from Hyalella montezuma (Hm), H. ‘azteca’ individuals in Montezuma Well (Hamw), Black River (Bla), Bubbling Springs (Bub), Crescent Lake (Cres), Marshall Lake (Marsh), Rainbow Lake (Rain), Unnamed Pond (Unp) and Comins Lake (Com). The haplotypes considered by Witt & Hebert (2000) are indicated by numbers which give the clade and haplotype (e.g. 1–3 refers to clade 1 haplotype 3). Bootstrap percentages (2000 pseudoreplicates) are given above nodes. Lines below taxon labels indicate cluster membership.
| Clust | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 0.025 ± 003 | 0.017 ± 005 | 0.037 ± 012 | 0.053 ± 015 | 0.050 ± 015 | 0.050 ± 014 | 0.078 ± 017 |
| 2 | 0.085 ± 010 | 0.041 ± 005 | 0.039 ± 011 | 0.055 ± 015 | 0.056 ± 014 | 0.055 ± 014 | 0.079 ± 016 |
| 3 | 0.232 ± 020 | 0.213 ± 020 | 0.018 ± 003 | 0.045 ± 014 | 0.030 ± 011 | 0.031 ± 011 | 0.067 ± 011 |
| 4 | 0.227 ± 021 | 0.221 ± 021 | 0.251 ± 024 | — | 0.043 | 0.044 ± 014 | 0.063 ± 015 |
| 5 | 0.231 ± 021 | 0.226 ± 021 | 0.231 ± 022 | 0.208 ± 021 | 0.006 ± 003 | 0.029 ± 011 | 0.077 ± 016 |
| 6 | 0.245 ± 021 | 0.240 ± 021 | 0.241 ± 022 | 0.224 ± 021 | 0.204 ± 020 | 0.019 ± 003 | 0.059 ± 014 |
| 7 | 0.275 ± 023 | 0.260 ± 021 | 0.251 ± 022 | 0.252 ± 022 | 0.266 ± 022 | 0.270 ± 022 | 0.069 ± 006 |
Among these 27 sequences, 278 nucleotide sites were variable (265 sites among the ingroup alone), and 243 were phylogenetically informative using cladistic criteria. The overall mean pairwise (± SE) transition/transversion ratio (Ts/Tv) for pairwise sequence comparisons was 2.94 ± 0.421. The mean base frequencies among the 27 sequences were A, 0.22; C, 0.19; G, 0.26; T, 0.33 and there was no evidence for heterogeneous nucleotide composition across the taxa (homogeneity χ2 = 44.75, d.f. = 78, P > 0.99).
The hierarchical likelihood ratio tests of 56 DNA sequence substitution models indicated the data were best explained by the HKY85 model (Hasegawa et al. 1985), with invariant sites and gamma distributed rate variation (HKY85 + I + Γ). The phylogeny estimated by ML with likelihood parameters estimated by modeltest (gamma distribution shape parameter α = 1.3624, proportion of invariant sites = 0.52, Ts/Tv = 4.67) suggests that clade 2, which includes H. montezuma, is paraphyletic with respect to clade 1 (Fig. 2). This topology is identical to the majority rule consensus of 9850 trees obtained in the Bayesian phylogenetic analysis (Fig. 2).

Maximum likelihood phylogram (–Ln likelihood = 4480.89). This topology is identical to the majority rule consensus of 9850 trees estimated in the Bayesian phylogenetic analysis. Numbers above nodes give the ML bootstrap percentages (500 pseudoreplicates), followed by the Bayesian posterior probabilities given as a percentage. The Bayesian posterior probabilities are equivalent to the bipartition frequencies among the 9850 trees, and give the node credibility given the data and model of nucleotide substitution.
To reduce the impact of homoplasy on phylogeny estimation, the maximum parsimony (MP) analysis was conducted with transversions and transitions weighted 3 : 1 on the basis of the overall pairwise mean Ts/Tv ratio. The g1 skewness statistic was highly significant (g1 = −0.55, g1crit = −0.37, P = 0.01) indicating nonrandomised phylogenetic signal in the data set. The MP analysis resulted in 53 equally parsimonious trees (length = 1314 steps, consistency index = 0.52, retention index = 0.80) (Fig. 3). Maximum parsimony converged on an estimate of phylogeny similar to the likelihood and Bayesian analyses, but the cladistic analysis suggests that clade 2 is monophyletic with moderate support, and also suggests clade 3 is sister to the branch terminating in clades 1 and 2. Support for the positions of most of the deep nodes was weak in these analyses.

Majority rule consensus cladogram of 53 equally parsimonious trees. Numbers above nodes give bipartition frequencies as a percentage among the 53 trees, followed by bootstrap percentages (2000 pseudoreplicates).
Discussion
This study has demonstrated that at least four (clades 1, 2, 6, 7) of the seven Hyalella species identified by Witt & Hebert (2000) occur in Arizona, and one of these (clade 2) is also present in Nevada. Individuals in Bubbling Springs Pond, although affiliated with clade 7, exhibit considerable sequence divergence from other members of this clade and likely represent another cryptic biological species, but for the purposes of this study are considered part of clade 7. Similarly, the population in the Black River is likely composed of two noninterbreeding morphologically cryptic taxa. This, however, must still be substantiated with reproductive compatibility trials in the laboratory, or the demonstration of differentiation at nuclear loci under sympatric conditions in nature. These results are consistent with those obtained by Thomas et al. (1997), who employed RAPDs and behavioural analysis to examine divergence among Hyalella populations in Arizona. The deepest genetic partition in their analysis was concordant with two behavioural groups: clingers and swimmers. However, additional genetic partitions were evident within these two groups, the shallowest of which occurred between H. montezuma and an H. ‘azteca’ population.
The phylogeny estimated by ML and Bayesian methods differed slightly from the phylogeny estimated by MP, and all three methods differed from that presented by Witt & Hebert (2000), who suggested that clade 7 was the sister group to the rest of the H. azteca complex. This difference can be explained by more appropriate character weighting, sequence evolution models, and a more suitable outgroup in the present study. Although the deep internal branching structure was not well resolved, H. montezuma clearly shares a common ancestry with members of clade 2, indicating that its morphological and ecological differentiation has been recent. Using strict phylogenetic species criteria, where species are designated on the basis of reciprocally monophyletic groupings, H. montezuma cannot be considered a distinct species because its evolution has been too recent for the establishment of reciprocal monophyly with respect to clade 2. In fact, ML and Bayesian analyses suggest clade 2 itself is paraphyletic with respect to clade 1, and MP does not strongly support the monophyly of clade 2. However, H. montezuma is so distinctive on the basis of ecological, biological and morphological criteria that downgrading of its taxonomic status is difficult to justify. Instead, the results of this study provide additional evidence for discordance between rates of molecular and morphological/ecological evolution within the genus Hyalella. In comparison to all other lineages within the H. azteca complex including members of clade 2, H. montezuma is highly mucronate (spinose), and has abnormally enlarged, highly setose mouth parts which facilitate filter feeding and its planktonic habit (Cole & Watkins 1977; Blinn & Johnson 1982; Wagner & Blinn 1987). Several cryptic small-bodied Hyalella species (clades 3, 4, 5, 6) exhibit up to 5 times as much sequence divergence as H. montezuma in comparison to members of clade 2, yet do not exhibit any clear morphological differences. Thus, the limited molecular divergence between H. montezuma and members of clade 2 demonstrates that evolutionary rate disparities are ‘two sided’ within the genus Hyalella, with deep genetic but limited morphological/ecological divergence, and limited genetic but strong morphological/ecological divergence patterns being evident.
A real time estimate for the divergence of H. montezuma is difficult to establish. Application of a standard COI clock rate of roughly 1.4% sequence divergence per million years (Knowlton & Weight 1998) suggests that H. montezuma diverged from other members of clade 2 approximately 3.8 million years ago. However, it is doubtful that the member of clade 2 with the closest phylogenetic affinity to H. montezuma (if it is extant) is represented in this study. In addition, current evidence suggests that several lineages within Hyalella have a very fast clock (J.D.S. Witt, in preparation), and that published calibrations may substantially overestimate divergence times. Given that Montezuma Well is thought to be about 120 000 years old (O.K. Davis, University of Arizona, personal communication), it would appear that the morphological and ecological differentiation of H. montezuma is likely to have occurred during this interval. What could account for the comparatively rapid morphological and ecological differentiation of this species in relation to other North American hyalellids?
Montezuma Well represents a unique point along the aquatic habitat continuum because its abnormally high CO2 concentration has prevented its colonization by fish, the absence of which has been linked to the evolution of H. montezuma (Cole & Watkins 1977). The planktonic habit of H. montezuma would render it highly susceptible to fish predation for two reasons. Firstly, fish are visual predators which exhibit size-biased prey selectivity, foraging on the largest prey available to them (Healy 1984; Wellborn et al. 1996). This category would include H. montezuma given that the species is substantially larger than cladocerans, copepods and other freshwater zooplanktors. Secondly, in comparison to their benthic counterparts, planktonic organisms have higher activity levels, which may be the most important attribute that enhances the susceptibility of a prey species to predators. As a result, most invertebrates that coexist with fish have lower activity levels than those in fishless habitats, or tradeoff body size against activity levels (Wellborn et al. 1996). As expected, H. montezuma has a much higher activity level than other Hyalella populations across Arizona (Blinn et al. 1988; Thomas et al. 1997).
Comparative ecological studies across a habitat gradient have provided important insights concerning the relationship between fish predation and ecological traits within the genus Hyalella. Wellborn (1994, 1995) examined Hyalella‘azteca’ populations across a gradient from fishless ponds to lakes inhabited by centrarchid fish in Michigan, USA. Fishless ponds were dominated by large-bodied species with larger adult body size, larger size at first reproduction and larger eggs than species that occurred in lakes with well developed fish communities. These traits were related to the different size-biased predation regimes experienced by the species; small body size decreases the predation risk imposed by fish predators, while large body size decreases the predation risk imposed by tactile invertebrate predators. Subsequent experiments have shown that these species also differ in activity levels, with the large species being more active (Wellborn 2002).
We hypothesize that fish predation is one factor that has constrained morphological and ecological evolution among North American hyalellid amphipods, and that its absence in Montezuma Well has permitted the rapid phenotypic differentiation of H. montezuma in comparison to other members of the the H. azteca complex. This study suggests that fish predation may not only impose selective constraints on body size and other life history traits (Wellborn 1994, 1995), but also on the ‘enemy free space’ (e.g. Jeffries & Lawton 1984) available for exploitation. A close examination of Hyalella in other large fishless habitats will likely reveal populations in the early stages of transition to a planktonic habit.
Cryptic speciation has been common among freshwater invertebrates, indicating that morphological stasis is an important motif in the evolution of freshwater life which requires explanation. The selective ecological factors involved in constraining morphological/ecological diversification among cryptic taxa are difficult to identify because they cannot be easily distinguished from developmental constraints (Schlichting & Pigliucci 1998). However, the results of this and other work (e.g. Wellborn 1994, 1995; Wellborn et al. 1996; Thomas et al. 1997; Witt & Hebert 2000) suggests that this problem is not intractable, and that comparative molecular and ecological studies across habitat gradients can provide important clues as to the selective ecological factors which constrain phenotypic diversification. Most past work has focused on examining the role of competition in determining ecological and morphological diversification (Schluter 2000), and studies investigating the role of predation regimes in the diversification of taxa are comparatively rare (but see Holt & Lawton 1994; McPeek 1995; McPeek & Brown 2000). In the case of Hyalella, ecological experimentation, which seeks to measure the response of morphological, life history and behavioural traits after long-term exposure to different predation regimes, promises important new insights into the role of predation in constraining phenotypic evolution.
Acknowledgements
We thank Doug Threloff for providing specimens from Nevada and Angela Hollis for aiding with the sequencing. Louis Bernatchez, Teri Crease, Melania Cristescu, Beren Robinson and anonymous reviewers provided helpful comments on earlier drafts of the manuscript. This study was supported by an Ontario Graduate Scholarship to J.D.S.W., Northern Arizona University Regents’ Professor funds to D.W.B., as well as NSERC and Canada Research Chairs program grants to P.D.N.H.
References
Jonathan Witt is currently working on his PhD, which is focused on determining the factors governing rates of morphological and molecular evolution among freshwater invertebrates, as well as the comparative biogeography and phylogeography of aquatic organisms. Dean Blinn's research focuses on the biology of extreme aquatic habitats, palaeolimnology, and aquatic population biology and genetics. Paul Hebert's research interests include molecular evolution, biodiversity and DNA based identification systems.




