Phylogeny, taxonomy, genetics and global heritage ranks of an imperilled, freshwater snail genus Lithasia (Pleuroceridae)
Abstract
Numerous aquatic species are threatened with extinction from habitat elimination or modification. One particularly imperilled group is the freshwater gastropod family Pleuroceridae. Pleurocerids reach their greatest diversity in the southeastern United States, and many species are currently considered extinct, endangered or threatened. One issue hindering efforts to implement conservation management plans for imperilled pleurocerid species is that the taxonomy is in an abysmal state. The taxonomy of pleurocerids is currently based on late 19th- and early 20th-century studies, which used a typological or morphospecies concept. Most biologists today doubt the validity of many of the currently recognized species; however, this does not stop them from assigning conservation ranks in an attempt to determine which species are imperilled or currently stable. We conducted a phylogenetic analysis of the pleurocerid genus Lithasia using morphological and mitochondrial DNA sequence (mtDNA) data in an attempt to delimit species boundaries and test previous taxonomic schemes. We found that the current taxonomy of Lithasia does not reflect species diversity adequately within the genus, with two new undescribed species being discovered. The conservation status ranks of the new, undescribed species are imperilled and would have been overlooked had we relied on the conventional taxonomy. Additionally, the undescribed species’ conservation ranks that were previously apparently secure became vulnerable due to being inappropriately assigned as members of formerly widely distributed species instead of the imperilled status they warrant and vice versa, as some taxa that were considered imperilled are now thought to be modestly stable. Our study suggests that conservation ranks should be considered suspect at best in taxonomically poorly known groups until the taxa are reviewed using modern systematic methods.
Introduction
The continued loss of aquatic species richness and diversity can be attributed to widespread anthropogenic elimination and degradation of freshwater habitats (Erwin 1991). This problem may be best illustrated by freshwater molluscs, which are in rapid decline thanks in part to river impoundment, habitat degradation, siltation, poor land use practices and the negative impact of introduced species (Stein 1976; Williams et al. 1993; Bogan et al. 1995; Lydeard et al. 1997). At least 70% of North American freshwater bivalves and 41% of freshwater gastropods are considered to be threatened, endangered or of special concern (Williams et al. 1993; Neves et al. 1997; Master et al. 1998). Attempts to preserve remaining taxa, therefore, must recognize unique processes and lineages so that they may be sustained in conservation efforts. This information can be used to assess and formulate conservation efforts for these groups (May 1990; Faith 1992; King et al. 1999). However, many freshwater mollusc groups present challenges to conservation biologists due to extensive taxonomic uncertainty, phenotypic plasticity, complex life histories and varied modes of reproduction (Bogan & Parmalee 1983; Kat 1983; Williams & Mulvey 1994; Lydeard et al. 1997; Lydeard & Roe 1998; Lydeard et al. 2000).
One group that exemplifies both a loss of diversity and a poor understanding of systematic relationships is the pleurocerid snails of North America. The family Pleuroceridae are gill-breathing, operculate snails found in North America and Asia, which reach their greatest diversity in the southeastern United States (Burch & Tottenham 1980; Lydeard & Mayden 1995; Lydeard et al. 1997). Of the 156 currently recognized pleurocerid species, at least 32 (including one genus, Gyrotoma) are considered extinct, and five are listed federally as endangered or threatened. The bulk of pleurocerid studies have focused on individual species and/or populations (e.g. Chambers 1978, 1980; Dillon 1984, 1988; Stiven & Kreiser 1994; Dillon & Lydeard 1998), and only recently have efforts been made to understand the systematics of the family and the species therein (Lydeard et al. 1997, 1998; Holznagel & Lydeard 2000; Lydeard et al. 2002). This lack of understanding regarding familial and generic composition hinders efforts to recognize, manage and conserve evolutionary significant units (Waples 1991; Mayden & Wood 1995) within the Pleuroceridae and other groups.
The genus Lithasia is one of six extant pleurocerid genera that are distributed throughout the Ohio, Cumberland and Tennessee river drainages (Burch & Tottenham 1980). Lithasia is comprised of 14 species and subspecies (Burch & Tottenham 1980) that have been recognized and classified differently by many authors. Most authors have used shell (Tryon 1873; Goodrich 1940; Burch 1982) or developmental (Davis 1974) characters in their classifications, placing the various species in different subspecific groups, subgenera, and genera. Little is known concerning the natural history of Lithasia, except that specimens are found generally on algae-covered rocks and logs (Conrad 1834; Tryon 1873; Davis 1974; Bogan & Parmalee 1983). The current conservation status of Lithasia varies from imperilled to apparently secure, with one species unknown and another possibly extinct (Table 1). Many Lithasia species have limited distributions, and one species, L. hubrichti, is presumed extinct (P. Hartfield pers. comm.). The systematics of Lithasia were addressed in Holznagel & Lydeard (2000), where they suggested the polyphyly of the genus and Leptoxis based on 16S rDNA sequences, while showing the monophyly of Elimia, Juga and Pleurocera.
| Taxon | Historical distribution | Rank | Proposed new name | Current distribution | New rank |
|---|---|---|---|---|---|
| armigera | Cumberland, Ohio, Tennessee River drainages (AL, IL, IN, KY and TN) | G3G4 | armigera | Cumberland, Ohio, Tennessee River drainages (AL, IL, IN, KY, TN) | G3 |
| curta | Tennessee River, AL | GU | ‘curta’ | Tennessee River, AL | GU |
| duttoniana | Duck River, TN | G2 | ‘fuliginosa’ | Duck River, TN | G3 |
| geniculata geniculata, geniculata fuliginosa | Cumberland, Duck, Tennessee River drainages (AL, TN) | G3G4 | ‘fuliginosa’geniculata’ new species | Duck River, TN Cumberland River and some upper Duck River tributaries, TN Buffalo River, TN | G3 G3 |
| geniculata pinguis | Collins River, Duck River headwaters, TN | G3G4 | ‘fuliginosa’ N/A | Duck River, TN Collins River, TN | G3 — |
| hubrichti | Big Black River, MS | GH | ‘hubrichti’ | Big Black River, MS | GX |
| jayana | Caney Fork and forks of Cumberland River, Duck River, TN | G2 | armigera | Cumberland, Ohio, Tennessee River drainages (AL, IL, IN, KY, TN) | GU |
| ‘fuliginosa’ | Duck River, TN | G3 | |||
| lima | Elk and Tennessee River drainages (AL, TN) | G2 | ‘lima’ | Elk River, Tennessee River and tributaries (AL, TN) | G3G4 |
| obovata | Green River drainage (KY); Ohio River drainage (IL, IN, KY) | G3G4 | N/A | Green River drainage, KY; Ohio River drainage (IL, IN, KY) | — |
| salebrosa salebrosa, salebrosa florentiana | Cumberland, Duck, Tennessee River drainages (AL, TN) | G3G4 | ‘fuliginosa’geniculata lima | Duck River, TN Cumberland River and some upper Duck River tributaries, TN Elk and Tennessee River drainages (AL, TN) | G3 G3 G3G4 |
| salebrosa subglobosa | Tennessee River, AL | G3G4 | ‘lima’ | Elk and Tennessee River drainages (AL, TN) | G3G4 |
| verrucosa | Mississippi, Ohio and Tennessee River drainages (AL, AR, IL, IN, KY, TN) | G3G4 | ‘lima’‘verrucosa’ new species | Elk and Tennessee River drainages (AL, TN) Ohio River drainage (IL, IN, KY) White River drainage, AR | G3G4 G3 G2G3 |
Proper species diagnoses can have a major impact on subsequent conservation strategies. Most freshwater mollusc species concepts, although often not clearly stated, can be classified as either morphological (Shull 1923; Regan 1926; du Rietz 1930) or taxonomic (Blackwelder 1967), relying on discrete or sometimes even ambiguous shell characters. The primary reference of currently recognized species of freshwater molluscs (Turgeon et al. 1998) is based largely on 19th- and early 20th-century taxonomy. Given the morphological variability presented by freshwater molluscs, and that morphological and taxonomic species concepts inherently exclude potentially valid species from definition (Mayden 1997), other methods must be used to accurately identify species. The use of the operational phylogenetic species concept (PSC) is historically based (Baum & Donoghue 1995) and includes the criterion of monophyly in the general sense (de Queiroz & Donoghue 1988) or ‘exclusivity’ (i.e. where an exclusive group of organisms is one whose members are related more closely to each other than they are to any organisms outside the group) to denote the recognition of species (Baum & Donoghue 1995). The PSC has helped clarify issues of conservation priority, species identity, group composition and diversity and the associated classification and taxonomy in these taxa (Mulvey et al. 1997; Roe & Lydeard 1998; Lydeard et al. 2000). We chose to employ the PSC as our operational method of taxon recognition, and tested the taxonomically and/or morphologically delineated species of Lithasia using mitochondrial DNA sequences employing the PSC (Mayden 1997; Lydeard et al. 2000). In addition, we examine how our PSC-based view of species alters the conservation status of the taxa.
Materials and methods
Specimen collection and sequencing
Specimens representing all recognized (Burch & Tottenham 1980) Lithasia species and subspecies were collected live from throughout their ranges where available (Fig. 1, Table 2) and stored frozen or in 95% ethanol. Specimen vouchers were deposited at the Illinois Natural History Survey as well as in the University of Alabama Gastropod Collection. L. hubrichti is considered extinct (P. Hartfield pers. comm.), and individuals bearing the L. curta and L. salebrosa subglobosa phenotypes were not found. Specimens of Io fluvialis and Leptoxis species were also collected and formed the ingroup with Lithasia. Three species of Pleurocera and Elimia hydei were chosen as outgroup taxa, as the two genera are sister to Io + Lithasia + Leptoxis (Holznagel & Lydeard 2000).
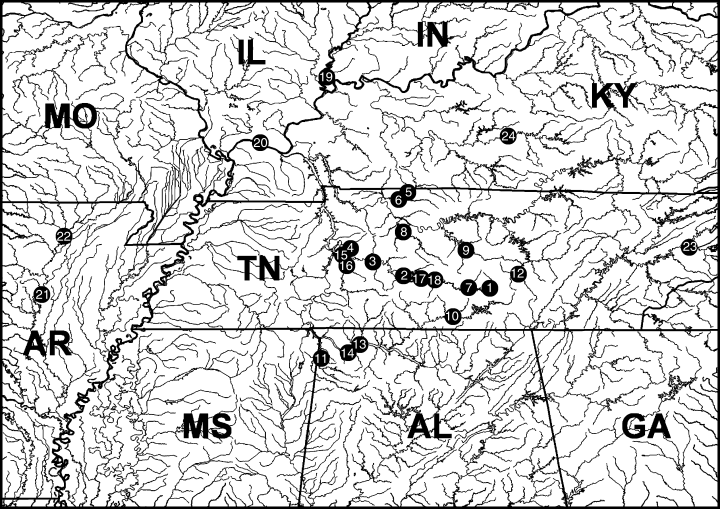
Map of collecting sites for Lithasia specimens. Locality and species information for each site is given in Table 2.
| Taxon | Identifier | Locality | n | Site | Accession no. | GenBank no. |
|---|---|---|---|---|---|---|
| Genus ‘Athearnia’ | ||||||
| A. anthonyi | A. anthonyi L. | Limestone Creek, Limestone Co., AL | 1 | UAG 581 | AF435772 | |
| A. anthonyi S. | Sequatchie R., Marion Co., TN | 1 | UAG 582 | AF435773 | ||
| A. anthonyi T. | Tennessee R., Jackson Co., AL | 1 | UAG 583 | AF435774 | ||
| Genus Elimia | ||||||
| E. hydei | E. hydei | Black Warrior R., Tuscaloosa Co., AL | 1 | UAG 584 | AF435775 | |
| Genus Io | ||||||
| Io fluvialis | Io C | Clinch R., Hancock Co., TN | 1 | UAG 585 | AF435776 | |
| Io N | Nolichucky R., Greene Co., TN | 1 | UAG 586 | AF435777 | ||
| Io P | Powell R., Lee Co., VA | 1 | UAG 587 | AF435778 | ||
| Genus Leptoxis | ||||||
| Le. praerosa | Le. praerosa H | Harpeth R., Davidson Co., TN | 1 | UAG 404 | AF435779 | |
| Le. praerosa Q | Sequatchie R., Marion Co., TN | 1 | UAG 588 | AF435781 | ||
| Le. praerosa S | Shoal Creek, Lawrence Co., TN | 1 | UAG 560 | AF435780 | ||
| Le. praerosa T | Tennessee R., Jackson Co., AL | 1 | UAG 589 | AF435782 | ||
| Genus Lithasia | ||||||
| Li. armigera | armigera H | Harpeth R., Cheatham Co., TN | 3 | 8 | UAG 555 | AF435739 |
| armigera O | Ohio R., Massac Co., IL | 2 | 20 | INHS 23628 | AF435740 | |
| armigera S | East Fork Stones R., Rutherford Co., TN | 6 | 9 | UAG 397 | AF435741 | |
| armigera T | Tennessee R., Lauderdale Co., AL | 1 | 14 | UAG 569 | AF435742 | |
| armigera W | Wabash R., White Co., IL | 2 | 19 | INHS 23634 | AF435743 | |
| Li. duttoniana | duttoniana 1 | Duck R., Maury Co., TN | 3 | 2 | UAG 402 | AF435744 |
| duttoniana 2 | Duck R., Maury Co., TN | 1 | 17 | UAG 578 | AF435745 | |
| Li. geniculata geniculata | geniculata geniculata | Duck R., Lillard Mill, Marshall Co., TN | 1 | 18 | UAG 580 | AF435755 |
| Li. geniculata fuliginosa | geniculata fuliginosa B1 | Buffalo R., Humphreys Co., TN | 1 | 15 | UAG 406 | AF435747 |
| geniculata fuliginosa B2 | Buffalo R., Perry Co., TN | 1 | 16 | UAG 395 | AF435748 | |
| geniculata fuliginosa D1 | Duck R., Maury Co., TN | 3 | 2 | UAG 403 | AF435749 | |
| geniculata fuliginosa D2 | Duck R., Maury Co., TN | 1 | 17 | UAG 579 | AF435750 | |
| geniculata fuliginosa D3 | Duck R., Hickman Co., TN | 3 | 3 | UAG 393 | AF435751 | |
| geniculata fuliginosa G | Garrison Fork, Bedford Co., TN | 1 | 7 | UAG 557 | AF435752 | |
| geniculata fuliginosa R1 | Red R., Robertson Co., TN | 1 | 5 | UAG 551 | AF435753 | |
| geniculata fuliginosa R2 | Red R., Robertson Co., TN | 6 | 6 | UAG 398 | AF435754 | |
| Li. geniculata pinguis | geniculata pinguis D1 & D2 | Duck R., Coffee Co., TN | 6 | 1 | UAG 392 | AF435763, −64 |
| geniculata pinguis C1 & C2 | Collins R., Warren Co., TN | 2 | 12 | UAG 407 | AF435761, −62 | |
| Li jayana | jayana | Duck R., Humphreys Co., TN | 1 | 4 | UAG 573 | AF435756 |
| Li lima | lima B | Bear Creek, Colbert Co., AL | 3 | 11 | UAG 570 | AF435757 |
| lima E | Elk R., Lincoln Co., AL | 3 | 10 | UAG 562 | AF435758 | |
| Li. obovata | obovata 1 & 2 | Green R., Hart Co., KY | 2 | 24 | UAG 574 | AF435759, −60 |
| Li. salebrosa salebrosa | salebrosa | Tennessee R., Lauderdale/Colbert Co., AL | 1 | 13 | UAG 565 | AF435765 |
| Li. salebrosa florentiana | florentiana | Tennessee R., Lauderdale/Colbert Co., AL | 3 | 13 | UAG 566 | AF435746 |
| Li. verrucosa | verrucosa B | Black R., Lawrence Co., AR | 2 | 22 | UAG 575 | AF435766 |
| verrucosa F | French Broad R., Knox Co., TN | 3 | 23 | UAG 576 | AF435767 | |
| verrucosa I | Wabash R., White Co., IL | 2 | 19 | INHS 23633 | AF435768 | |
| verrucosa O | Ohio R., Massac Co., IL | 2 | 20 | INHS 23629 | AF435769 | |
| verrucosa T | Tennessee R., Lauderdale/Colbert Co., AL | 3 | 13 | UAG 568 | AF435770 | |
| verrucosa W | White R., Woodruff Co., AR | 2 | 21 | UAG 577 | AF435771 | |
| Genus Pleurocera | ||||||
| P. canaliculatum | P. canaliculatum | Duck R., Maury Co., TN | 1 | UAG 590 | AF435783 | |
| P. prasinatum | P. prasinatum | Coosa R., Shelby Co., AL | 1 | UAG 591 | AF435784 | |
| P. walkeri | P. walkeri | Shoal Creek, Lauderdale Co., AL | 1 | UAG 592 | AF435785 | |
- INHS = Illinois Natural History Survey; UAG = University of Alabama Gastropod Collection.
Genomic DNA was isolated from head tissues using standard phenol/choloroform methods. Mitochondrial DNA sequences were obtained for an amplified segment of the mitochondrial cytochrome oxidase c subunit I gene using primers GASCOIH (5′-TTTAT-TACTACTATTTTTAATATACG) and GASCOIL (5′-GGAATATTATTATCTCA-TTCTAAAGC) designed from consensus sequences of Littorina saxatilis (Wilding et al. 1999) and Katharina tunicata (Boore & Brown 1994). The primers amplify an approximately 1000 base pairs (bp) fragment of the gene and were used for amplification and sequencing. Double-stranded amplifications via PCR were generated using 50–500 ng of template genomic DNA in 25 µl volumes (10 mm Tris, 50 mm KCl, 2.5 mm MgCl2, 1 µm each primer, 0.1 mm each dNTP, 1.5 units Taq DNA polymerase; Fisher Scientific). The amplification regime consisted of: initial denaturation (92 °C for 120 s); 30 cycles of denaturation (92 °C for 40 s); annealing (40 °C for 40 s) and extension (72 °C for 90 s); and a final extension (72 °C for 180 s). After the first five cycles, the annealing temperature was raised to 50 °C. Double-stranded products were concentrated using Millipore Ultrafree MC filters and provided the template for cycle sequencing using the ABI BigDye kit following manufacturer's instructions. Reactions were purified using Qiagen DyeEx spin columns and sequenced on an ABI 3100 genetic analyser.
Sequence analyses
Sequences were entered in the software program xesee (version 3.0; Cabot & Beckenbach 1989) and aligned by eye with the aid of L. saxatilis sequence. The aligned matrix is available from the authors. To account for the possibility of multiple COI haplotypes within a single population, additional individuals from each population, when available, were sequenced and included in the analysis. Sample sizes are shown in Table 2.
Because any analytical method, including parsimony, tends to group sequences of similar nucleotide composition together regardless of evolutionary history (Lockhart et al. 1994), a χ2 test of base frequency homogeneity was run in paup* (version 4.0b10; Swofford 2002) to test for composition bias. Sequences were examined for evidence of saturation by plotting the number of transitions and transversions at each codon position against uncorrected p-distance using mega 2.0 (Kumar et al. 2001). Skewness of tree-length distributions as a measure of phylogenetic signal (Hillis & Huelsenbeck 1992) was estimated by generating 10 000 random trees in paup*. To account for genetic variation within and among species and genera, uncorrected p-distances between all samples were calculated.
Phylogenetic hypotheses were generated under parsimony and maximum likelihood. For the parsimony analysis, 50 replicates of heuristic search with random addition were run in paup* with the following settings: random sequence addition, tree-bisection–reconnection (TBR) branch swapping and branches with minimum length equal to zero collapsed. To test the internal stability of the data, a jackknife analysis (Farris et al. 1996; 1000 replicates, 10 random additions per replicate) was performed using xac (Farris unpublished) and Bremer support values (1000 trees; Bremer 1994) were generated in nona 2.0 (Goloboff 1998). For the ML analysis, the matrix was analysed using modeltest 3.06 (Posada & Crandall 1998) to determine an appropriate model of evolution. The data were then analysed in paup* using the same settings as the parsimony analysis, with empirical nucleotide frequencies used and remaining values estimated under the TrN + G model (Tamura & Nei 1993) with six categories of substitution and a gamma distribution.
Molecules and morphology
Because most classifications of Lithasia are based on morphological features (Tryon 1873; Goodrich 1940; Burch & Tottenham 1980), 25 shell and radula characters were coded (Minton 2002) and added to the molecular data. Minton (2002) showed that these characters roughly define pleurocerid genera and some previously considered species groups (Fig. 2). The resulting matrix was analysed under maximum parsimony in paup*, again using Pleurocera and Elimia taxa as outgroups and jackknife support determined as before using pac (Farris unpublished) with the same options as in the parsimony sequence analysis.
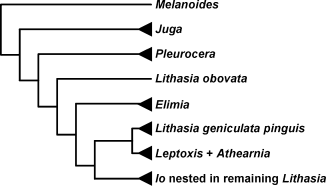
Modified phylogeny of Minton (2002) based on 25 shell and radula characters.
Biogeography
In addition to noting gross patterns of geographical division in the molecular analyses, we constructed area cladograms based on distribution and phylogeny of Lithasia only and compared them to speciation hypotheses to look for any replicated geographical or faunal distribution patterns. Using the method of Wiley (1988), river drainages were viewed as ‘taxa’, and the taxa occurring in those drainages as ‘characters’ (see Crandall & Templeton 1999). River drainage designations followed Mayden (1988), with two exceptions. Garrison Creek was considered a unique drainage; while it is part of the upper Duck River drainage, it is considered to have a Cumberlandian faunal distribution (Ortmann 1924; van der Schalie 1973; Ahlstedt 1980). The lower Ohio River was also considered a distinct drainage, as the area where included taxa occur does not appear in any of the predetermined drainages. Using the strict consensus phylogeny generated by total evidence analysis, terminal taxa were coded for what drainage they occurred in, and interior nodes were coded for those taxa that were in the clade and their distributions (Wiley 1988). The resulting matrix (Table 3) was analysed using paup*.
| 1 | 2 | 3 | 4 | 1 | 2 | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | |||
| Lower Tennessee | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| Upper Tennessee | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Ohio | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| Wabash | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| White | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Black | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Cumberland | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 |
| Garrison Creek | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Buffalo | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| Duck | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| Mobile | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Results
Molecular analyses
Aligned sequences with no necessary gaps, representing 47 total and 23 Lithasia haplotypes resulted in a data matrix of 890 characters, of which 306 were variable and 257 were parsimony informative (aligned matrix available from authors). A χ2 test of base frequency homogeneity showed no composition bias (χ2 = 27.64, d.f. = 138, P = 1.0000). While most sequence diversity occurred at the third codon position, transition/transversion plots showed no evidence of saturation at any codon position (data not shown). Random tree length distribution indicated significant phylogenetic signal in the data (g1 = −0.4722).
Recent studies (e.g. Sunnucks & Hales 1996; Zhang & Hewitt 1996; Lovette et al. 1999; Williams & Knowlton 2001) have reported the presence of nuclear copies of mitochondrial genes (‘pseudogenes’/‘numts’) that may be amplified and sequenced, thereby confounding phylogenetic analyses. We believe that the sequences reported here are mitochondrial in origin, based on a single amplicon after PCR, ‘clean’ sequencing reads of both strands, a lack of internal stop codons or frameshifts and base frequency and substitution patterns comparable to other pleurocerid studies. The existence of or changes in any of the above may indicate the presence of nuclear sequence (Collura & Stewart 1995; Sunnucks & Hales 1996; Sorenson & Quinn 1998).
Maximum parsimony analysis yielded 15 trees (766 steps, CI = 0.52) rendering Lithasia nonmonophyletic (Fig. 3). Specimens of L. obovata formed a clade between Elimia and Pleurocera species, while two specimens of L. geniculata pinguis from the Collins River fell between Elimia hydei and the Leptoxis clade. The remaining Lithasia species formed a clade sister to Io. Within the ‘Lithasia’ clade, two major clades were recovered: one containing specimens of L. lima, L. salebrosa and its subspecies and L. verrucosa; and one comprised of L. armigera, L. duttoniana, L. geniculata and its subspecies, and L. jayana. Within the L. verrucosa clade, three subclades were recovered: one containing specimens from the White River drainage of Arkansas; one containing lower Ohio River specimens; and one of Tennessee River drainage specimens. The position of Tennessee River verrucosa could not be determined in relation to L. lima and L. salebrosa and its subspecies. Within the second clade, two subclades were recovered. All L. armigera specimens grouped together sister to a clade of L. geniculata fuliginosa from the Cumberland and upper Duck rivers. The second subclade showed L. g. fuliginosa specimens from the Buffalo River sister to a clade containing taxa from the lower Duck River. This lower Duck River clade contained specimens of L. geniculata and its subspecies along with L. duttoniana and L. jayana. L. g. pinguis specimens in this clade possessed unique haplotypes, as did L. jayana, while the remaining L. geniculata and L. duttoniana shared a single haplotype. The relationships within the clade could not be resolved. Many currently recognized taxa were not recovered as monophyletic. Only L. armigera was recovered in all trees. L. geniculata and L. verrucosa were rendered polyphyletic, and the remaining taxa were unresolved.
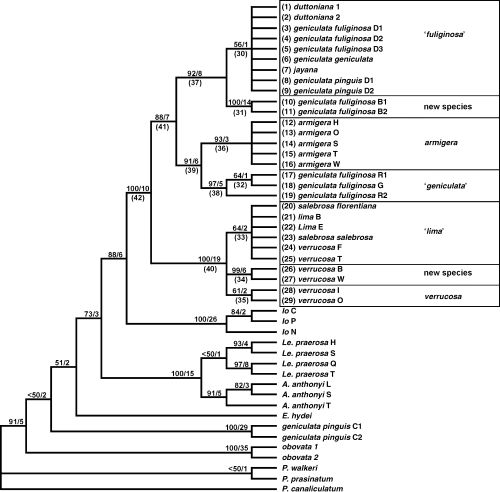
Strict consensus of 15 equally most parsimonious trees (766 steps, CI = 0.52) generated using sequence data. Taxa identified as in Table 2. Numbers above each node are jackknife values > 50%/B remer support values. Numbers before taxa and under certain nodes correspond to character number used in biogeography analysis. Proposed species identifications are outlined. Names in quotes are suggested for that taxon but not fixed in this study.
Maximum likelihood analysis yielded a single tree (Fig. 4). Lithasia was rendered nonmonophyletic, with E. hydei, L. geniculata pinguis C1 and C2 and L. obovata 1 and 2 forming a clade. The remaining Lithasia species formed a single clade with a topology consistent with that achieved under parsimony.

Single tree generated under maximum likelihood using sequence data. Taxa identified as in Table 2.
Genetic variation within and among groups
Of all the populations sampled in the ‘Lithasia’ clade, only L. g. pinguis from the Duck River showed the presence of multiple haplotypes, differing by an uncorrected p-distance of 0.45%. Genetic distances for all groups are shown in Table 4. The maximum interpopulation difference within a putative species was 4.27% between L. g. fuliginosa from the Duck and Red Rivers, while many pairs (e.g. L. verrucosa B and W, O and I, etc.) showed no difference. The maximum distance seen within the ‘Lithasia’ clade was 9.10% between L. jayana and L. s. salebrosa. Within-genus distances were less than between genus distances in all cases except for the ‘Elimia’ clade and between Athearnia and Leptoxis.
| Genus | Max. distance within | Distance between | ||||
|---|---|---|---|---|---|---|
| Athearnia | ‘Elimia’ | Io | Leptoxis | ‘Lithasia’ | ||
| Athearnia | 3.03% | |||||
| ‘Elimia’ | 11.91% | 11.35–12.25% | ||||
| Io | 0.45% | 11.24–11.68% | 11.01–12.70% | |||
| Leptoxis | 7.88% | 4.83–7.21% | 11.46–14.41% | 12.25–13.61% | ||
| ‘Lithasia’ | 9.10% | 11.46–13.16% | 12.47–15.52% | 9.66–11.93% | 11.91–14.98% | |
| Pleurocera | 6.74% | 10.23–11.68% | 8.20–11.91% | 9.89–10.02% | 10.34–13.06% | 10.67–14.06% |
Molecules and morphology
Analysis of the combined data sets yielded 12 trees of 872 steps (CI = 0.47; Fig. 5). Lithasia was again rendered polyphyletic by the positions of L. g. pinguis from the Collins River and L. obovata. The overall topology was consistent with that generated using only molecular data, although a number of clades (e.g. L. verrucosa specimens and L. armigera) were better resolved. Jackknife support was similar to that generated solely using molecular data.
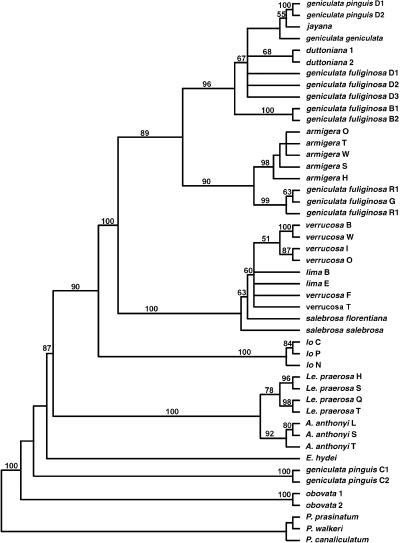
Strict consensus of 12 equally parsimonious trees (872 steps, CI = 0.47) generated in a combined analysis of sequence and morphological data. Taxa identified as in Table 2.
Biogeography analysis
Clear biogeographical divisions were seen in the analyses. Using molecular data, all remaining Lithasia taxa formed clades based on river drainages with the exception of L. armigera. The subclades containing L. verrucosa were divided into Ohio, White and Tennessee river drainages, although the positions of each within the larger clade could not be resolved. Lithasia geniculata taxa were divided into Buffalo, Cumberland/upper Duck and lower Duck river groups. All lower Duck River taxa formed a single clade. Jackknife analysis strongly supported the division of Lithasia taxa into their respective clades.
Parsimony analysis of area relationships produced two trees (57 steps, CI = 0.91; Fig. 6). Two major clades were resolved: Cumberland/Duck river drainages; and a Tennessee, lower Ohio and Ozark (Black and White rivers) drainages. This pattern is inconsistent with previously generated analyses (e.g. Mayden 1988; Crandall & Templeton 1999), as the ‘lower Ohio’ (Cumberland, Duck, lower and upper Tennessee) and Interior Highland rivers (Wabash, lower Ohio, Black and White) did not group together. The pattern is also inconsistent with other speciation hypotheses in North American rivers (Mayden 1988).

The two equally parsimonious trees (57 steps, CI = 0.91) generated in biogeography analysis of data in Table 3. See text for explanation of procedures and character coding.
Discussion
Species composition of Lithasia
Although historically considered a genus based on morphological characters, Lithasia was rendered polyphyletic in all analyses, with L. obovata and L. g. pinguis from the Collins River falling outside the clade of all remaining species. Burch (1982) noted that based on shell characters, L. obovata and L. g. pinguis appear to belong to Elimia/Pleurocera and Leptoxis, respectively, but are placed in Lithasia because of their radulae. This hypothesis was supported by Minton (2002), who showed that L. obovata fell between Elimia and Pleurocera, and L. g. pinguis grouped with Leptoxis using shell and radula characters. Based on mitochondrial DNA sequences alone and combined with morphological data, L. obovata again fell between Pleurocera and Elimia taxa, suggesting that it is not a Lithasia species and needs to be studied further for proper taxonomic placement. Taxa believed to be L. g. pinguis were present in two parts of the trees. Specimens from the Duck River grouped with other Duck River taxa in the main ‘Lithasia’ clade, while two specimens from the Collins River each possessed unique haplotypes and grouped with Elimia species. L. g. pinguis is known from the Caney Fork and its branches and the Duck River, Coffee County, TN (Burch & Tottenham 1980). These two populations represent at least two separate taxa, and only those from the Duck River should be considered Lithasia. Again, further work needs to be undertaken to determine the identity and placement of the Collins River taxa. The remaining taxa form a well-supported clade that should be recognized as the genus Lithasia.
Genetic variability within Lithasia
Genetic divergences seen in Lithasia, as defined above, were comparable to literature values for mitochondrial 16S rDNA sequence variation in pleurocerids (Lydeard et al. 1997, 1998; Holznagel & Lydeard 2000). The maximum difference within a putative species was 4.27% for L. g. fuliginosa, exceeding the largest published value of 3.7% at 16S for populations of Elimia carinocostata (Lydeard et al. 1998). The maximum intrageneric difference within Lithasia was 9.10%, well within the 8.69% and 10.1% reported by Holznagel & Lydeard (2000) at 16S for Pleurocera and Elimia, respectively, and below the 12–15% they reported for Leptoxis. Overlapping intra- and intergeneric divergences were seen in ‘Elimia’ and between Athearnia and Leptoxis. Athearnia is frequently considered congeneric with Leptoxis (Burch & Tottenham 1980; Turgeon et al. 1998; Holznagel & Lydeard 2000; but see Morrison 1971; Dillon & Ahlstedt 1997), and similar patterns of overlap were reported for Elimia (Lydeard et al. 1998; Holzangel & Lydeard 2000).
Species delineation
Several versions of the PSC recognize species as minimal monophyletic groups (de Queiroz & Donoghue 1988; Mishler & Donoghue 1982; Mishler & Brandon 1987). This has proved useful in a variety of studies on freshwater bivalves (Mulvey et al. 1997; Roe & Lydeard 1998; King et al. 1999; Lydeard et al. 2000) and gastropods (Lydeard et al. 1998). The PSC further provides a framework for recognizing unique monophyletic lineages based on sequence divergence when morphological distinctness historically used to define species is problematic (e.g. Wilke & Falinowski 2000). This is useful in pleurocerids, where cryptic species that are not closely related have been observed (Chambers 1978). Using this concept, seven phylogenetic species (Fig. 3) would be recognized based on the molecular phylogeny of Lithasia (3, 4): (1) L. ‘fuliginosa’, a composite of what were formerly L. duttoniana, L. g. fuliginosa, L. g. geniculata, L. jayana and L. g. pinguis, all of which are from the Duck River; (2) a new, previously undescribed species comprised of L. g. fuliginosa from the Buffalo River; (3) L. armigera, the only currently recognized morphospecies recovered as monophyletic; (4) L. ‘geniculata’, a group comprised of L. g. fuliginosa from the upper Duck River tributaries and Cumberland River drainage; (5) L. lima, a composite comprised of L. lima, L. verrucosa from the Elk and Tennessee rivers, L. s. salebrosa and L. s. florentiana; (6) a new, previously undescribed species comprised of L. verrucosa from the White River drainage, Arkansas; and (7) L. ‘verrucosa’, comprised of L. verrucosa from the Ohio River drainage. Based on the combined morphological and molecular data and biogeography analyses, it is possible that L. jayana and L. salebrosa (salebrosa and florentiana) may warrant recognition as morphologically diagnosable, phylogenetic species. However, we await the acquisition of additional genetic data to examine this further.
The molecular and combined molecular + morphology data analyses uncovered hidden diversity within what were once considered widely distributed, highly variable morphospecies. Many of the newly recognized phylogenetic species have distributions that correspond quite well with geography (Fig. 6). The distinction between Cumberlandian and upper Duck river fauna from lower Duck River mollusc fauna has been noted by other authors. In their works on freshwater mussels, Ortmann (1924) and van der Schalie (1973) both noted the upper Duck River contains Cumberlandian species while the lower Duck River contains species of the Ohioan/Interior faunal group and those of undetermined origin (Ahlstedt 1980). The sister relationships of the Duck and Buffalo river taxa is not surprising, as the Buffalo River is the main tributary to the Duck River and they share faunal assemblages (Ortmann 1924; van der Schalie 1939, 1973). Lithasia ‘lima’, the undescribed sp. from the White River, and L. ‘verrucosa’ showed similar geographical division. The undescribed sp. is from the Black, Spring and White river drainages of Arkansas, L. ‘lima’ is from widely distributed in the Tennessee River and L. ‘verrucosa’ is from the Wabash and lower Ohio rivers (Goodrich 1940; Burch & Tottenham 1980). This general pattern of separation is consistent with freshwater fish and crayfish distributions and river drainages (Crandall & Templeton 1999; Mayden 1985, 1988).
Conservation implications from a PSC–based classification
Employing the PSC considerably modified the traditional classification of Lithasia, resulting in the synonymization of some subspecies and species and recognition of newly delineated species. The Natural Heritage Network has developed a method for estimating the degree of relative imperilment (Master et al. 2000). Global conservation status ranks are based on a one-to-five scale, ranging from critically imperilled (G1) to widespread, abundant and demonstrably secure (G5). With the new phylogenetically based classification, however, the conservation status has been altered appreciably. For example some taxa, such as L. lima, which is now part of the phylogenetic ‘lima’ has gone from G2 to G3G4, while some subspecies now constitute more narrowly restricted phylogenetic species, making them rarer (e.g. two new undescribed species have gone from G3G4 to G2; Table 1). The findings of this study suggest that other widely distributed pleurocerid taxa in particular (e.g. Elimia livescens, Leptoxis praerosa, Pleurocera canaliculatum and P. vestitum), and poorly delineated taxonomic groups in general, may actually be comprised of several phylogenetic species which warrant protection at a different level than currently recognized.
Acknowledgements
We thank S. Ahlstedt, K. Cummings, J. Garner, T. Watters and other colleagues for providing specimens for study. A. Bogan, P. Churchill, K. Cummings, R. Dillon, R. Mayden, P. J. West and anonymous reviewers provided helpful comments on the manuscript. Financial support was provided by the Conchologists of America (RLM), the US Fish and Wildlife Service (CL) and the National Science Foundation (CL).




