Phylogeny and ecological radiation of New World thistles (Cirsium, Cardueae – Compositae) based on ITS and ETS rDNA sequence data
Abstract
Sequence data from a portion of the external transcribed spacer (ETS) and internal transcribed spacers (ITS-1 and ITS-2) of 18S-26S nuclear ribosomal DNA were used to resolve historical biogeography and ecology of true thistles (Cirsium, Cardueae, Compositae) in the New World. The 650 base-pair, 3′ portion of the ETS examined here showed a level of variation across taxa similar to that of the ITS sequences included. A maximum-likelihood tree based on combined ETS and ITS sequences leads us to suggest that the New World species of true thistles constitute a major lineage, which in turn comprises several smaller lineages. A western North American lineage shows weak quartet-puzzling support, but includes a well-supported lineage of species endemic to the California Floristic Province. Comparisons of this Californian lineage with other neoendemic angiosperm groups of the region show that the Californian Cirsium lineage exhibits unusually high ecological diversity for a group displaying such low levels of rDNA sequence divergence across taxa. Similarly low levels of sequence divergence were found throughout the New World Cirsium lineage. These results indicate either that Cirsium underwent a rapid ecological radiation in North America, or that rDNA evolution in North American Cirsium has been highly conservative.
Introduction
The concept of adaptive radiation has played an important role in the development of evolutionary biology. The term was coined by Henry Fairfield Osborn (1902), but notable examples, such as the Galápagos finches (Geospizidae), have inspired evolutionary biologists from the time of Darwin (1859). Despite these well-known examples, different workers have defined adaptive radiation in various ways (for a discussion, see Givnish 1997). In general, definitions of adaptive radiation either have emphasized an increase in diversification rate, sometimes after acquisition of a putative ‘key innovation’, or ecological shifts within a lineage (see Schluter 2000).
Under any definition of adaptive radiation, the key factors are only meaningful in comparisons across lineages. For example, a rapid increase in diversification rate can be detected by estimating the timing of branching events across a phylogenetic tree or examining relative diversity of sister-lineages (see Sanderson & Donoghue 1996). Alternatively, comparisons may be implied rather than explicit. Osborn (1902), in citing the rise of Australian marsupials as an example of adaptive radiation, was comparing their ecological and morphological diversity to that of an inferred, most recent common ancestor. In his example, the length of time since divergence was not of central importance to defining this event as an adaptive radiation.
Assuming a single colonization event, New World true thistles (Cirsium) may qualify as an example of adaptive radiation from the standpoint of overall ecological diversity across lineages. To satisfy a more stringent definition of adaptive radiation, demonstration that this radiation happened relatively rapidly, compared to other lineages of North American plants, is necessary. In this study of the New World Cirsium we contrast ecological and genetic diversity in the group in an effort to gain insight into the timing of New World thistle diversification. We use phylogenetic analysis of 18S–26S nuclear rDNA transcribed spacers [external transcribed spacer (ETS) and internal transcribed spacer (ITS)] to identify a lineage of New World Cirsium that can be compared to other angiosperm lineages of similar maximum age and geographical distribution. In particular, we have chosen to examine several lineages that have evolved during the climatic innovation of summer drought within the California Floristic Province.
A minority of lineages of Cardueae are indigenous to the New World. The true thistles (Cirsium) are the most diverse of these native lineages, with approximately 80 indigenous species of perennial to biennial herbs in North America (Ownbey et al. 1975). Although many of these species are well delimited, taxa in some species groups (e.g. the C. occidentale complex and some species of the Great Plains) are poorly defined taxonomically due to introgression or incipient speciation (David J. Keil, pers. comm.). Other New World lineages of Cardueae are relatively depauperate, i.e. Saussurea (approximately three species in northwestern North America), Centaurea (two species in the USA and approximately three species in South America), and Centaurodendron (three species endemic to the Juan Fernandez Archipelago).
Many native North American species of Cirsium are characterized by populations with few individuals and many taxa are narrow endemics. For example, of 28 native taxa recognized by Keil & Turner (1993) in The Jepson Manual: Higher Plants of California as occurring in California, 10 are listed in the California Native Plant Society's Inventory of Rare and Endangered Plants of California as globally rare or endangered (CNPS 2001). Under the US Endangered Species Act, Cirsium hydrophilum var. hydrophilum, C. loncholepis, C. fontinale var. fontinale and C. fontinale var. obispoense are classified as endangered and C. pitcheri and C. vinaceum are listed as threatened. Some native North American species of Cirsium, however, have thrived in areas of human disturbance (e.g. C. discolor) and a small number have become troublesome weeds in some areas (e.g. C. undulatum, C. ochrocentrum).
North American members of Cirsium are found in a great variety of plant communities, such as prairies (e.g. C. hillii and C. carolinianum), montane meadows (e.g. C. scariosum and C. subniveum), rocky desert canyons (e.g. C. arizonicum and C. neomexicanum), seeps and streamsides (e.g. C. fontinale and C. crassicaule), brackish marshes (e.g. C. hydrophilum var. hydrophilum and C. loncholepis), forest (e.g. C. brevistylum and C. remotifolium), coastal dunes (e.g. C. rhothophilum), lacustrine dunes (e.g. C. pitcheri) and openings in chaparral (e.g. C. occidentale var. venustum). The vast majority of North American species of Cirsium are restricted to specific plant communities. Taxa that occur in multiple plant communities (e.g. C. occidentale sensulato and the C. scariosum complex) are morphologically diverse, taxonomically difficult and may be undergoing active diversification (Keil & Turner 1993; Keil, personal communication).
Petrak (1917) published the most recent infrageneric classification of North American Cirsium more than 80 years ago. Petrak classified all North American taxa in Cirsium subg. Eucirsium. The subgenus, in turn, was divided into six North American sections, three of which are monotypic. The three sections comprising more than one species include various subsections and series. Groups ranked at the level of series represent Petrak's interpretation of closely related species groups. Some of these groupings have been called into question in later taxonomic treatments. For example, C. walkerianum (Petr.) J.T. Howell (sect. Onotrophe subsect. Acanthophylla) was recognized as a taxonomic synonym of C. quercetorum (A. Gray) Jeps. (sect. Onotrophe subsect. Acaulia) by Keil & Turner (1993).
Given the lack of understanding of relationships within North American Cirsium, the group is a prime candidate for phylogenetic analysis utilizing DNA sequence data. Within angiosperms, the internal transcribed spacer (ITS) region of 18S–26S nuclear ribosomal DNA has provided phylogenetic signals within many, putatively young groups of congeners, including a wide diversity of lineages in Compositae (see Baldwin et al. 1995; Soltis & Soltis 1998). Susanna et al. (1995) and Garcia-Jacas et al. (2001) found that ITS sequences provide evidence of relationships among species groups within the thistle tribe (Cardueae), in subtribe Centaureinae. The external transcribed spacer (ETS) upstream from the 18S region of 18S–26S nuclear ribosomal DNA also has been used in phylogenetic studies of various tribes of Compositae (e.g. Baldwin & Markos 1998; Clevinger & Panero 2000; Linder et al. 2000; Chan et al. 2001; Markos & Baldwin 2001; Lee et al. 2002). In Compositae, the 3′ portion of the ETS region, located c. 400–650 base pairs (bp) upstream of the 18S gene, is at least as variable as ITS-1 and ITS-2 combined and has proved to be an excellent adjunct to phylogenetic data from the ITS region (e.g. Baldwin & Markos 1998; Clevinger & Panero 2000; Linder et al. 2000; Chan et al. 2001; Markos & Baldwin 2001; Lee et al. 2002). However, no highly conserved region exists within close proximity to the 5′ end (transcription initiation site) of the ETS (see Volkov et al. 1996) and an internal primer must be designed. ITS and ETS sequences are the most rapidly evolving DNA regions that have been used for studying phylogeny in a broad range of plant groups.
In this study, we estimate phylogeny of the New World thistles using maximum-likelihood analysis of ITS and ETS rDNA sequences. Using these results, we identify a lineage from the California Floristic Province (Howell 1957; Raven & Axelrod 1978) that provides a contrast in ecological and genetic (rDNA) diversity with other neoendemic angiosperm lineages of the region, but not with the rest of the New World Cirsium lineage.
Materials and methods
Fifty-two plants representing 35 native North American Cirsium taxa, 10 Old World Cirsium taxa, and two species each of Carduus and Onopordum were included in the study (Table 1). Taxa were chosen to represent a broad sample of diversity, especially within New World Cirsium, as revealed from literature and herbarium specimens. Sampled Old World taxa include species of tribe Cardueae naturalized in North America, as well as European and Asian species that have not invaded the New World. Tissue was obtained from recently collected herbarium specimens (less than 20 years old) or from fresh leaf material and seedlings. Sampled seedlings (except those that did not survive) were grown to maturity for vouchering. Voucher specimens were deposited at the University Herbarium (UC), University of California, Berkeley. DNA was extracted in CTAB buffer using 0.1–0.3 g of leaf tissue following the procedure of Doyle & Doyle (1987).
| Taxon | Voucher/source | GenBank no. | |
|---|---|---|---|
| ITS | ETS | ||
| Carduus nutans (Petr) Arènes | Seed ex CDFA | AF443678 | AF443730 |
| Carduus tenuiflorus Curtis | Crockett, CA; DGK 01.025 | AF443679 | AF443731 |
| Cirsium andersonii (A. Gray) Petr | Seed ex CDFA | AF443678 | AF443735 |
| Cirsium andrewsii A. Gray | San Francisco Co, CA; DGK 01.027 | AF443684 | AF443736 |
| Cirsium arvense (L.) Scop. EUR | Seed ex France | AF443681 | AF443733 |
| Cirsium arvense IL | Cook, Co, IL | AF443680 | AF443732 |
| Cirsium arvense MT | Gallatin Co, MT; DGK 99.040 | AF443682 | AF443734 |
| Cirsium brevistylum Cronquist | Sonoma Co, CA; DGK 01.028 | AF443685 | AF443737 |
| Cirsium calcareum (Jones) Wooton & Standl. | San Juan Co, UT; RDS 2307 | AF443687 | AF443739 |
| Cirsium canovirens Rydb. | Siskiyou Co, CA; CAS 640931 | AF443688 | AF443740 |
| Cirsium canum M. Bieb. | Seed ex hort; DGK 01.030 | AF443689 | AF443741 |
| Cirsium congdonii R. J. Moore & Frankton | Mono Co, CA; RDS 23489 | AF443690 | AF443742 |
| Cirsium cymosum (Greene) Jeps. | Seed ex CDFA | AF443691 | AF443743 |
| Cirsium discolor (Willd) Spreng. | Hamilton Co, OH; DGK 97.017 | AF443692 | AF443744 |
| Cirsium douglasii DC. | Sonoma Co, CA; DGK 01.029 | AF443686 | AF443738 |
| Cirsium eatonii B.L. Rob. | Box Elder Co, UT; RDS 1999 | AF443694 | AF443746 |
| Cirsium edule | Mt Ranier, WA; DGK 035 | AF443711 | AF443763 |
| Cirsium ehrenbergii Sch. Bip. | Durango, Mex; UC1611642 | AF443726 | AF443778 |
| Cirsium fontinale Greene | Seed ex San Mateo, CA | AF443695 | AF443747 |
| Cirsium f. var. obispoense J.T. Howell | San Luis Obispo Co, CA; DGK 01.032 | AF443696 | AF443748 |
| Cirsium faucium Petr. | Mexico, MexUC 1476531 | AF443725 | AF443777 |
| Cirsium hydrophilum (Greene) Jeps. | Seed ex Solano Co, CA | AF443698 | AF443750 |
| Cirsium henryi (Franch.) Diels | Hubei, China; UC 149 0827 | AF443697 | AF443749 |
| Cirsium jorullense Spreng. | Hidalgo, Mex; UC 1519954 | AF443699 | AF443751 |
| Cirsium lineare Sch. Bip. | Hubei, China; UC 1490729 | AF443727 | AF443779 |
| Cirsium lomatolepis Petr. | Mex, Mex; UC 172380 | AF443724 | AF443776 |
| Cirsium mohavense (Greene) Petr. | Inyo Co, CA; RDS 1006 | AF443700 | AF443752 |
| Cirsium monocephalum Lév. | Hunan, China; CAS 916686 | AF443701 | AF443753 |
| Cirsium monspessulanum Hill. | Seed ex ARC | AF443717 | AF443769 |
| Cirsium muticum Mich. | UC 1354709 | AF443722 | AF4437 |
| Cirsium neomexicanum A. Gray | Maricopa Co, AZ; CAS 945400 | AF443718 | AF443770 |
| Cirsium occidentale (Nutt.) Jeps. | Marin Co, CA | AF443702 | AF443754 |
| Cirsium o. var. venustum (Greene) Jeps. | Inyo Co, CA; RDS 2333; DGK 01.044 | AF443703 | AF443755 |
| Cirsium palustre (L.) Scop. | Seed ex Bavaria, Germany; DGK 01.033 | AF443704 | AF443756 |
| Cirsium pitcheri (Eaton) Torr. & Gray | Seed ex ARC | AF443705 | AF443757 |
| Cirsium quercetorum (A. Gray) Jeps. | Solano Co, CA; DGK 01.034 | AF443706 | AF443758 |
| Cirsium remotifolium (Hook.) DC. | Linn Co, OR; CAS 964060 | AF443707 | AF443759 |
| Cirsium rhaphilepis Petr. | Guanajuato, Mex; UC 1502312 | AF443708 | AF443760 |
| Cirsium rhothophilum S. F. Blake | Seed ex CDFA | AF443709 | AF443761 |
| Cirsium rydbergii Petr. | San Juan Co, UT; CAS 881549 | AF443710 | AF443762 |
| Cirsium scariosum Nutt. | Seed ex ARC | AF443693 | AF443745 |
| Cirsium spinosissimum Scop. | Seed ex Plannersee, Austria | AF443720 | AF443772 |
| Cirsium subniveum Rydb. | Cash Co, UT; RDS 1969 | AF443712 | AF443764 |
| Cirsium tioganum Petr. | BCN, Mex; UC 1523693 | AF443721 | AF443773 |
| Cirsium tymphaeum Hausskn. | Seed ex Grakenland, Denmark; DGK 01.037 | AF443723 | AF443775 |
| Cirsium utahense Petr. | Box Elder Co, UT; RDS 2170 | AF443713 | AF443765 |
| Cirsium velatum Petr. | Jalisco, Mex; UC 1612991 | AF443714 | AF443766 |
| Cirsium vulgare (Sav.) Ten. CA | Contra Costa Co, CA; DGk 01.036 | AF443715 | AF443767 |
| Cirsium vulgare EUR | Seed ex Bas-Rhin, France; DGK 01.038 | AF443716 | AF443768 |
| Cirsium wheeleri Petr. | Garfield Co, UT; CAS 560803 | AF443719 | AF443771 |
| Onopordum acaulon L. | Ex hort | AF443676 | AF443728 |
| Onopordum cf. illyricum | Sicily, Italy; DGK 97.013 | AF443677 | AF443729 |
- UC = University Herbarium, Berkeley; CDFA = California Department of Food and Agriculture, Sacramento, CA; CAS = California Academy of Sciences, San Francisco, CA; DGK = Dean G. Kelch; RDS = R. Douglas Stone.
ETS primer design
Primers 18S-ETS (ACT TAC ACA TGC ATG GCT TAA) (Baldwin & Markos 1998) and ETS-Car-1 (TTC GTA TCG TTC GGT) were used for amplifying and sequencing the 3′ portion of the ETS examined in our study. Primer ETS-Car-1 was designed by sequencing upstream from the 18S gene into the IGS of Carduus nutans and Cirsium andrewsii and locating a region of conserved sequence in the two taxa, c. 660 bp from the ETS/18S gene border. The IGS of these taxa were amplified using primers 18S–IGS and 26S–IGS (Baldwin & Markos 1998). IGS polymerase chain reactions (PCR) comprised 25 µL of 50 mm Tris-HCL, 2.5 mm MgCl2, 16 mm (NH4)2SO4, 150 µg/mL bovine serum albumin, 250 mm of each dNTP, 1 µm of each primer, 0.25 µL of Klentaq-LA by Clonetech and approximately 1–10 ng of template DNA. IGS PCR was conducted in thin-walled microfuge tubes using an MJ Research PTC-200 Peltier thermal cycler with the following parameters: initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s and 68 °C for 7 min. Superior results were obtained by using the above machine rather than machines with slower temperature ramping rates.
ITS and ETS sequencing
Primers ITS4 (White et al. 1990) and ITS-I (Urbatsch et al. 2000) were used to amplify the ITS region (ITS-1, 5.8S gene and ITS-2) and primers 18S-ETS and ETS-Car-1 were used to amplify the 3′ end of the ETS via the PCR, following the protocol of Baldwin & Wessa (2000). For short-distance PCR of the ITS and the 3′ end of ETS, we used 25 µL reaction volumes including 1.25 µL glycerol, 50 mm KCl, 10 mm Tris-HCL, 2.5 mm MgCl2, 250 µm of each dNTP, 0.5 µm of each primer, 0.5 units of AmpliTaq DNA Polymerase (Perkin-Elmer) and 1–10 ng of genomic DNA. PCR was conducted in thin-walled tubes in an MJ Research PTC-200 Peltier thermal cycler thermal cycler at 97 °C for 1 min (initial denaturation), followed by 40 cycles of 97 °C for 10 s, 48 °C (for ITS) or 55 °C (for ETS) for 30 s (annealing stage), and 72 °C for 1 min (extension phase), and concluding with 72 °C for 7 min (final extension stage).
Resulting products were cleaned using Millipore Ultrafree-MC tubes. Cycle sequencing reactions for the ITS PCR products were conducted using a Big Dye terminator cycle sequencing premix kit (Perkin-Elmer). Four µL of sequencing reagent premix, 0.4 µm of primer ITS-4 or ITS-5 (White et al. 1990) for ITS or primer 18S-ETS or ETS-Car-1 for ETS, 2 µL of DMSO and 20–40 ng of cleaned PCR products were added to thin-walled microfuge tubes. Cycle sequencing was performed in an MJ Research PTC-200 Peltier thermal cycler with the following settings: initial denaturation at 97 °C for 1 min, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s and 60 °C for 4 min. Sequencing products were cleaned using isopropanol precipitation, then resolved on a 4.8% Long-Ranger polyacrylamide gel using an Applied Biosystems 377 automated sequencer (Perkin-Elmer).
Data analysis
Sequence data from the ITS and ETS were edited and easily aligned manually using Sequence Navigator, version 1.0.1. Gaps were coded as missing data. Approximately 1.4% of the matrix cells are represented as missing data. Copies of the aligned data set can be obtained from Treebase (SN1124). DNA sequences for all taxa in this study have been deposited in GenBank (see Table 1). Sequence data were compiled into a nexus file and analysed phylogenetically using maximum likelihood, as implemented in paup* test version 4.0d64 for Macintosh PPC (Swofford 2001). Phylogenetic analyses were conducted using the HKY85 model of sequence evolution and the heuristic algorithm of paup (with as-is stepwise addition and TBR branch swapping). Quartet-puzzling values were calculated to assess branch support in the resulting trees (Strimmer & von Haeseler 1996). All New World taxa were investigated for rate constancy of ETS and ITS sequence evolution using a likelihood ratio test for differences between clock-constrained and clock-unconstrained trees (Felsenstein 1988). The maximum likelihood analyses were conducted using the HKY85 model of sequence evolution, a gamma distribution of rate variation among sites (with the shape parameter set to four), and no assumed invariant sites. The difference in the likelihood values was examined for significance using a χ2 test with the degrees of freedom equal to n-2 (n= number of terminal branches).
Average genetic divergence values were calculated for the endemic Californian Cirsium lineage, as well as for the Compositae lineages Baeriinae and Madiinae (Baldwin & Wessa 2000), Stephanomeria (Lee et al. 2002), Sidalcea (Andreasen & Baldwin 2001), Sanicula sect. Sanicoria (Vargas et al. 1999), the perennial lineage of Eriophyllum (Baldwin unpublished data) and Lessingia (Markos & Baldwin 2001). Clades originating in the California Floristic Province (CFP) were chosen using character optimization methods in MacClade (Maddison & Maddison 1999). When circumscription of CFP clades was equivocal, then a conservative circumscription was adopted by choosing the smaller of potential clades. Corrected pairwise distances were obtained using the HKY85 model. To estimate average divergence within each lineage, pairwise distance values were averaged for pairs of taxa spanning the ingroup root node. Taxa dispersed across a tree were chosen for pairwise comparisons. To compute an average divergence value, trees with highly asymmetrical levels of diversity on either side of the ingroup root-node required fewer pairwise comparisons than did trees with symmetrical levels of diversity across the root-node. Because, due to shared ancestry, pairwise comparisons of taxa are not independent, no standard measures of variance could be computed for the resulting mean values.
Ecological diversity within each of the Californian lineages examined was estimated by summing the number of plant communities in which members of a lineage naturally occur. Plant community definitions were based on those of Holland & Keil (1995) and were judged to represent a particular combination of environmental values, such as insolation, soil chemistry, temperature range, moisture availability and fire history. Occurrence of species within a particular community was based on our field observations, as well as statewide and local floras (Mason 1957; Munz 1973, 1974; Thomas 1961; Sharsmith 1982; McClintock et al. 1990; Hickman 1993; Twisselmann 1967; Best et al. 1996; Matthews 1997; Smith 1998).
Results
Successful amplification of Cardueae IGS was achieved by using a thermocycler with rapid temperature ramping rates. Although three to several bands were obtained for each taxon, ranging in size from 1.8 to 4 kb in length, readable sequences were obtained for two taxa (Carduus nutans and Cirsium andrewsii) by means of direct sequencing. Successful direct sequencing from variously sized products may be the result of using a nested primer (all but one product class being the result of nonspecific amplification) or of IGS length variation occurring upstream from the sequenced region (see Baldwin & Markos 1998). A 21-bp sequence, invariant between the two sequenced taxa, was used to design primer ETS-Car-1 (Fig. 1), beginning 561 bp upstream from the 3′ end of ETS (referring to the sequence of Cirsium andrewsii). A subsequent PCR of 10 Cardueae taxa using ETS-Car-1 and 18S-ETS produced products of nearly identical length. Alignment of the ETS sequences was unambiguous for the Cardueae taxa sampled.
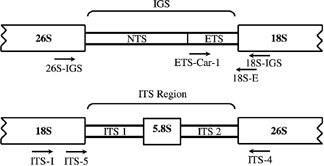
Relative location of primers used in this study. 18S = 18S ribosomal DNA; 26S = 26S ribosomal DNA; 5.8S = 5.8S ribosomal DNA. IGS = intergenic spacer; NTS = nontranscribed spacer (possible occurrence of an ETS at 26S end not shown); ETS = external transcribed spacer. Primer ETS-Car-1 begins 561 base pairs upstream from the 5′ end of the 18S gene.
Most ETS variation is between the ingroup and the outgroup or between members of the outgroup. Maximum pairwise sequence divergence for the ETS region examined is 14.8%, between Atractylodes japonica and Centaurea calcitrapa. Within Cirsium, the maximum pairwise sequence divergence for length variation of the ETS region examined was 5.2%, between C. arvense and C. henryi.
Alignment of the 52 ITS-region sequences of Cardueae yielded a matrix of 710 characters, 1–251 in ITS-1, 252–415 in 5.8S and 416–710 in ITS-2. Length of the ITS region varies from 686 bp in Carduus tenuiflorus to 692 bp in C. nutans.
As in the ETS, most variation in the ITS region is between the ingroup and the outgroup or between outgroup taxa. Among sampled taxa, the maximum pairwise sequence divergence for the ITS region is 13.9%, between Onopordum acaulon and O. sp. from Segesta, Sicily. Among sampled members of Cirsium, the maximum pairwise sequence divergence for the ITS region is 6.7%, between C. monocephalum and C. arvense.
Phylogenetic analysis of combined ITS and ETS data using maximum likelihood resulted in the tree shown in Fig. 2. Low ITS and ETS variation among members of Cirsium prevents robust assessment of any incongruent signal across data sets, but quartet-puzzling branch scores are higher for trees derived from combined data than for trees based on ITS or ETS data alone. Support for monophyly of Cirsium in relation to sampled species of other genera (Onopordum and Carduus) is moderately high, with a quartet puzzling value of 86. Quartet-puzzling support for monophyly of New World species (Cirsium subg. Eucirsium) is low (48), but this lineage is resolved in the maximum-likelihood tree (Fig. 2). Western North American taxa occur in several lineages with low to moderate quartet-puzzling support. A lineage of predominantly northwestern North American distribution comprises taxa with chromosome numbers of 2n = 34 (a number widespread in Cirsium; Moore & Frankton 1962; Ownbey 1968); an endemic Californian lineage includes most taxa with a chromosome number of 2n = 32 (Ownbey et al. 1975). Rate constancy of rDNA evolution among New World taxa is rejected.
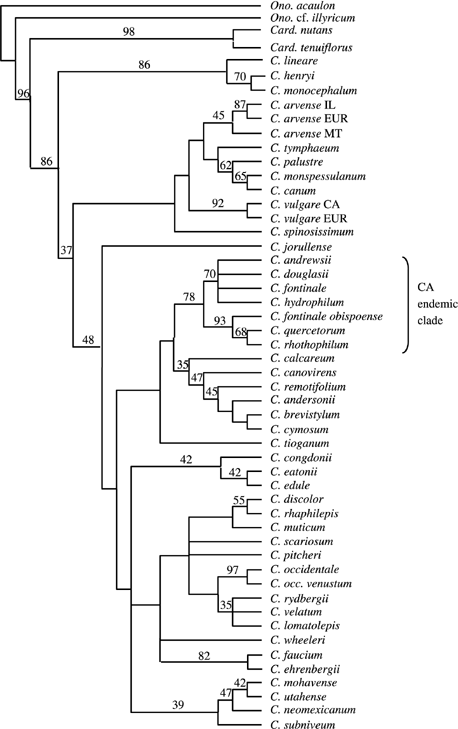
Maximum likelihood tree from analysis of the combined ITS and ETS sequence data. Numbers above branches represent quartet puzzling values over 34. Card. = Carduus; Ono. = Onopordum; C. =Cirsium; MT = Montana; IL = Illinois; CA = California; EUR = Europe; C. occ. = Cirsium occidentale.
A comparison of average pairwise divergence in ITS and ETS sequences among various principally Californian plant lineages indicates relatively low values for Cirsium. Average pairwise divergence in other groups range from 0.015 in Lessingia to 0.186 in Madiinae; average divergence within the endemic Californian Cirsium lineage is 0.010. In contrast, ecological divergence in the endemic Californian Cirsium lineage, as estimated by plant community occurrence, is relatively high in comparison to other plant groups examined (Table 2; Fig. 3).
| Taxon | Ci | Ba | Ma | Si | St | Sa | Le |
|---|---|---|---|---|---|---|---|
| Intertidal | |||||||
| Estuarine | |||||||
| Salt marsh | X | X | X | X | |||
| Dune | X | X | |||||
| Scrub dune | X | X | X | X | X | ||
| Dune wetland | X | X | X | ||||
| N coast scrub | X | X | X | X | |||
| S coast scrub | X | X | X | X | |||
| S semidesert coast. scrub | |||||||
| Sea-bluff | X | X | X | X | X | X | X |
| Mixed chaparral | X | X | X | X | X | X | |
| Chamize chaparral | X | X | |||||
| Red-shank chaparral | |||||||
| Manzanita chaparral | |||||||
| Ceanothus chaparral | |||||||
| Scrub oak chaparral | X | ||||||
| Maritime chaparral | X | X | X | ||||
| Island chaparral | X | ||||||
| Serpentine chaparral | X | X | |||||
| Montane chaparral | |||||||
| Semidesert chaparral | X | ||||||
| Native bunchgrass | X | X | X | X | X | X | X |
| Valley grassland | X | X | X | X | |||
| N coastal prairie | X | X | X | X | X | X | |
| Desert grasslands | X | X | X | X | |||
| Coast cypress/pine | X | ||||||
| Knobcone pine | |||||||
| Interior cypress | X | ||||||
| N coast conifer | X | ||||||
| Coast redwood | |||||||
| N mix evergreen | X | ||||||
| S mix evergreen | X | X | |||||
| Sierran hardwood | |||||||
| Coast live oak | X | X | X | X | |||
| Valley oak woodland | X | X | |||||
| Foothill woodland | X | X | X | ||||
| N oak woodland | X | X | X | ||||
| S oak woodland | X | X | X | X | X | ||
| Island oak woodland | X | ||||||
| Mixed conifer forest | X | X | X | X | X | X | |
| Red fir forest | |||||||
| Lodgepole pine forest | |||||||
| Subalpine forest | |||||||
| Alpine meadows | X | ||||||
| Rocky alpine meadow | X | X | |||||
| Desert alpine meadow | X | ||||||
| Pinon juniper woodland | X | X | X | X | |||
| Joshua tree woodland | X | X | |||||
| GB sagebrush scrub | X | ||||||
| Saltbush scrub | |||||||
| Blackbush scrub | X | ||||||
| Creosote bush scrub | X | X | X | X | |||
| Desert sand dune | X | ||||||
| Desert dry wash | X | X | X | ||||
| Alkali sink | X | ||||||
| W Valley desert scrub | X | ||||||
| Valley riparian | X | ||||||
| Mt riparian | |||||||
| Limnetic plants | |||||||
| Freshwater marsh | X | X | X | ||||
| Bog and fen | X | ||||||
| Montane meadow | X | X | |||||
| Vernal pool | X | ||||||
| Ruderal | X | X | |||||
| Number of communities | 18 | 27 | 26 | 10 | 18 | 18 | 11 |
| Number of species | 7 | 42 | 89 | 19 | 13 | 14 | 14 |
- Ci = California endemic clade of Cirsium (see Fig. 2); Ba = Baeriinae; Ma = Madiinae; Si = Sidalcea; St = Stephanomeria; Sa = Sanicula sect. Sanicoria; Le =Lessingia. Plant community classification follows Holland & Keil (1995). Total number of Californian species in each clade is listed at the bottom.

Ecological diversity versus genetic divergence. Horizontal axis represents the number of California plant communities where members of the clade are present following the system of Holland and Keil (1995). Vertical axis represnts the average pairwise distance (using the HKY 85 model) between taxa across the root node based on ITS and ETS rDNA sequence data (only ITS data are available for Sanicula sect. Sanicornia).
Discussion
ETS and ITS sequences in Cardueae
The ETS primers 18S-ETS and ETS-Car-1 are probably universal for the thistle tribe (Cardueae, Compositae) based on high yield of PCR products of uniform size (c. 650 bp) for taxa of Cardueae that span major lineages of the group. Absence of evident length variation in the 3′ region of the ETS (hereafter, ETS) of Cardueae contrasts with considerable length variation in the same region of another group of Compositae, Heliantheae sensu lato (Baldwin & Markos 1998). Use of the ETS for higher-level phylogenetic studies in Cardueae may be possible based on unambiguous alignment of the sequences across the disparate genera sampled. Length uniformity and ease of alignment of the ETS sequences may be indicative of an overall low level of variation in rDNA transcribed spacer sequences of the thistle tribe.
As in some other groups of angiosperms investigated to date [e.g. Sidalcea (Malvaceae), Andreasen & Baldwin 2001; Lasthenia (Compositae), Chan et al. 2001; Stephanomeria (Compositae), Lee et al. 2002)], the ETS and ITS-1 + ITS-2 appear to have evolved at similar rates in Cardueae. Average pairwise distances across the root node separating Onopordum acaulon from the other taxa are 0.10 for ETS and 0.15 for ITS-1 + ITS-2. Enhancement of resolution and support in rDNA trees by combined, rather than separate, analysis of ETS and ITS data accords with other studies indicating that the ETS often nicely complements phylogenetic data from the ITS region (e.g. Baldwin & Markos 1998; Clevinger & Panero 2000; Linder et al. 2000; Chan et al. 2001; Markos & Baldwin 2001; Lee et al. 2002).
Relationships among species
Use of rDNA transcribed spacers for resolving phylogeny within Cirsium is limited by low sequence variation among species but evidently not by the rate of concerted evolution. Conspecific populations of each of the three species represented by multiple samples (C. arvense, three samples; C. occidentale, two samples and C. vulgare, two samples) constitute lineages in the maximum-likelihood (ML) tree (Fig. 2). rDNA transcribed spacers in Cirsium appear to have undergone sufficiently rapid concerted evolution for use in phylogeographical study of at least one widespread species, C. arvense (Kelch, unpublished).
Strong support (quartet puzzling value = 86) for monophyly of Cirsium relative to the outgroup species sampled provides provisional evidence for the value of a pappus character for diagnosing the genus. Carduus is morphologically similar to Cirsium and differs most conspicuously from Cirsium by having nonplumose, rather than plumose, pappus bristles. Pappus evolution is known to have been highly dynamic in some groups of Compositae (e.g. in Heliantheae s.l., Baldwin et al. 2002), so reliance on a single pappus character for circumscribing genera of thistles might be regarded as questionable. Further sampling within Carduus is necessary to discern whether the genus and Cirsium are each truly monophyletic.
Putative monophyly of New World Cirsium (supported by a low quartet-puzzling value of 48; Fig. 2) leads us to suggest provisionally that Cirsium invaded North America only once before the advent of European peoples. If New World taxa of Cirsium indeed constitute a lineage, then indigenous North American members of the genus are each equally closely related to weedy thistles introduced into North America from the Old World. A single origin for New World Cirsium contradicts the traditional taxonomy, which treats New World species in multiple sections that each include Old World species (De Candolle 1837; Petrak 1917). For example, sect. Onotrophe includes the New World species C. crassicaule, C. edule and C. quercetorum, as well as the Old World species C. monspessulanum, C. palustre and C. spinosissimum. These taxa do not constitute a lineage in the ML tree (Fig. 2).
Within the New World thistles, one well-supported lineage (quartet puzzling score of 78) consists of species that are confined to the California Floristic Province (CFP; as delimited by Howell 1957; Raven & Axelrod 1978). This group contains rare endemics, including three of the four taxa of Cirsium that are listed as endangered by the US Fish and Wildlife Service. If a weakly supported lineage (quartet puzzling value of 16) consisting of the Californian endemic clade and a lineage of taxa from northwestern North America (some of which enter California in the Coast Ranges or Sierra Nevada) (Fig. 2) is upheld by future evidence, then all endemics of the CFP could be concluded to stem from the same western North American radiation (although probably from more than one ancestral source within the CFP).
The phylogeny presented here does not support the monotypic sections that Petrak (1917) erected for Californian taxa. The two monotypic sections sampled in our study, sect. Dermatolepis (C. fontinale) and sect. Mastigophyllum (C. rhothophilum), are both strongly nested within the lineage endemic to the CFP (Fig. 2). Petrak's taxonomy implies that C. fontinale and C. rhothophilum are isolated relicts; our results show that the two taxa are neoendemics, rather than palaeoendemics.
Ecological diversity in thistles of the California Floristic Province (CFP)
Like other regions on earth with a Mediterranean climate, the CFP has unusually high plant diversity and endemism for a temperate, continental area (Dallman 1998). Most plant groups restricted to the CFP are neoendemics that are believed to have evolved concurrently with or subsequent to the development of summer drought (Raven & Axelrod 1978; Axelrod 1992). Fossil and palaeoclimatic evidence indicate that the summer drying trend in western North America began at mid-Miocene (c. 15 Ma; e.g. Axelrod 1992; Flower & Kennett 1994).
Members of Cirsium that naturally occur within the CFP represent at least three separate lineages and cannot be reliably treated as representing a single example of radiation within the region (Fig. 2). Cirsium occidentale constitutes a highly variable lineage that occurs widely in western North America (including the CFP) and may be undergoing incipient diversification with zones of secondary introgression (Keil & Turner 1993; David Keil, pers. comm.). The Pacific Northwest lineage includes species that extend south into California along the coast (e.g. C. brevistylum) or along the higher mountain chains (e.g. C. andersonii). The California Endemic lineage includes various narrow endemics, most of which have a chromosome number of 2n = 32 (e.g. C. fontinale and C. rhothophilum; Ownbey et al. 1975). Only the California Endemic lineage was used in ecological comparisons. All taxa in the group are strongly associated with the CFP and therefore are likely to be products of a common radiation in the region that postdated the advent of the late Tertiary summer-drying trend (Axelrod 1992; Flower & Kennett 1994).
For purposes of comparison with Californian Cirsium, a conservative approach was used to identify other neoendemic angiosperm lineages of the CFP. For example, a clade corresponding to tribe Madieae (Compositae) sensu Baldwin comprises taxa (e.g. subtribe Baeriinae and subtribe Madiinae) that are mainly confined to the CFP (the Hawaiian silversword alliance being the primary exception) and others (e.g. Arnica) that are more widespread (Baldwin & Wessa 2000). Baeriinae, Madiinae and Arnica may represent a common CFP radiation or conceivably may represent separate colonizations of the CFP by different western North American ancestors. As with Cirsium, we chose the most cautious interpretation and treated Baeriinae and Madiinae as separate CFP radiations (Fig. 3). The number of species in each of these CFP endemic clades varied from seven in the California endemic clade of Cirsium to 89 in Madiinae, not including the Hawaiian taxa (Table 2). We chose mean genetic divergence to reflect divergence over time rather than taxon number, as the latter can be strongly skewed by artefacts of taxonomic judgement and extinction within lineages.
Ecological variation within the lineages compared in Fig. 3 is distributed mainly between, rather than within, taxa and appears to be largely the result of evolutionary shifts in ecology rather than phenotypic plasticity. In the California Endemic clade of Cirsium, taxa are narrowly restricted in habitat occurrence. In the other angiosperm groups, most taxa are confined to a limited number of plant communities, with notable exceptions (e.g. some perennial members of Eriophyllum and ruderal tarweeds).
Within the CFP, most angiosperm groups for which we have comparable rDNA and habitat data display a similar relationship between the level of rDNA divergence within a lineage and ecological breadth of the group (Fig. 3). The California Endemic lineage of Cirsium displays an extraordinarily high ecological diversity relative to the degree of rDNA divergence among taxa (Fig. 3). Unusually rapid ecological radiation or unusually low rates of ETS and ITS evolution could explain the results for Cirsium. Minimal chloroplast DNA divergence among the taxa of Cirsium in trnK intron/matK sequences (Kelch, unpublished) leads us to suspect that molecular divergence within the group is low in general and that rapid ecological radiation has occurred in the Californian thistles. Differences in rates of molecular evolution associated with differences in habit or life history (Gaut et al. 1996; Andreasen & Baldwin 2001) are unlikely to explain low rates of molecular evolution in the biennial or short-lived perennials in the California Endemic lineage of Cirsium; other angiosperm lineages included in our comparisons represent both annual and perennial groups.
High ecological diversity and low rDNA variation appear to extend throughout New World Cirsium and may reflect rapid ecological radiation on a continental scale. As noted above, other indigenous groups of Cardueae in North America are relatively depauperate and do not provide parallel examples of diversification in the New World. Columbines (Aquilegia, Ranunculaceae), on the other hand, constitute a particularly prominent and well-demonstrated example of extensive radiation of north temperate perennial herbs, with substantial diversity in North America (Hodges & Arnold 1994). In columbines, evolutionary radiation was mediated by changes in floral morphology and pollinator preference (Hodges 1997). In Cirsium, some morphological differentiation within and between species may be attributable to pollinator preference (e.g. flower colour and flower head length) but data on pollination and mating system are lacking. In general, differences in ecological setting or habitat among taxa of Cirsium are more pronounced than floral differences, as is evident in various examples of insular adaptive radiation in Compositae such as Hawaiian Madiinae (Baldwin 1997; Baldwin & Sanderson 1998), Macaronesian Sonchus (Kim et al. 1996; Kim et al. 1997) and Dendroseris (Sang et al. 1994; Esselman et al. 2000). In general, our data illustrates that Cirsium represents a continental parallel to insular composite lineages displaying low rDNA divergence and extensive ecological diversity.
Acknowledgements
We thank the Wyoming Biocontrol Weed Group, the USDA National Biological Control Institute and the Lawrence R. Heckard Fund of the Jepson Herbarium at UC Berkeley for providing funding for this research. Michael Sanderson and two anonymous reviewers provided suggestions that materially improved the manuscript. The California Department of Food and Agriculture, ARC Vegaville, the University of California Botanical Garden, Cindy Roché and Andre Gassmann generously provided seed. We also thank the University Herbarium, the California Academy of Sciences and R. Douglas Stone for access to herbarium material. Special thanks to Katarina Andreasen, Raymond Chan, L. D. Gottlieb, Joongku Lee, Staci Markos and Pablo Vargas for access to information on sequence divergence in California neoendemic groups. We are grateful to Fred Hrusa for providing information on taxon occurrences.
References
Dr Dean G. Kelch is a Research Associate at the University and Jepson Herbaria at the University of California. His research interests concern the phylogeny, biogeography and evolution within lineages of seed plants containing both relicts and neoendemics. Dr Bruce G. Baldwin is Curator of the Jepson Herbarium and Associate Professor in the Department of Integrative Biology, University of California. His research focuses on the evolution and phylogeny of young angiosperm lineages, particularly in western North America and on oceanic islands.




