Confirmation of low genetic diversity and multiple breeding females in a social group of Eurasian badgers from microsatellite and field data
Abstract
The Eurasian badger (Meles meles) is a facultatively social carnivore that shows only rudimentary co-operative behaviour and a poorly defined social hierarchy. Behavioural evidence and limited genetic data have suggested that more than one female may breed in a social group. We combine pregnancy detection by ultrasound and microsatellite locus scores from a well-studied badger population from Wytham Woods, Oxfordshire, UK, to demonstrate that multiple females reproduce within a social group. We found that at least three of seven potential mothers reproduced in a group that contained 11 reproductive age females and nine offspring. Twelve primers showed variability across the species range and only five of these were variable in Wytham. The microsatellites showed a reduced repeat number, a significantly higher number of nonperfect repeats, and moderate heterozygosity levels in Wytham. The high frequency of imperfect repeats and demographic phenomena might be responsible for the reduced levels of variability observed in the badger.
Introduction
In areas typified by agricultural landscapes in lowland England, the Eurasian badger (Meles meles) has an unusual social behaviour where groups of up to 25 animals defend a common territory, share a sett, allogroom, play and may show limited alloparental care (Neal 1986; Kruuk 1989; Woodroffe 1993). However, they forage, dig, travel and spend most of the time in solitary outside their setts (Kruuk 1989). The adaptive significance of badger facultative sociality is not fully understood (see reviews by Woodroffe & Macdonald 1993 and Johnson et al. 2000).
Badger groups include several mature males and females and are generally formed by recruitment of the offspring into their natal territory (Woodroffe et al. 1993). Litter size averages 2.4–2.7 across Europe (da Silva et al. 1994) and there is a high reproductive failure in social badgers compared to solitary badgers. This reproductive failure has been linked to group living and high population densities (Cresswell et al. 1992; Woodroffe & Macdonald 1995a). Reproductive suppression seems to be the result of female competition for resources, and there is little evidence that it is linked to co-operative care of the young (Woodroffe & Macdonald 1995a) as is common in other carnivores (Creel & Creel 1991; Creel & Macdonald 1995).
Several approaches, including behavioural observation (Cheeseman et al. 1987; Woodroffe & Macdonald 1995a), allozyme data (Evans et al. 1989; da Silva et al. 1994) and ultrasound scanning (Woodroffe 1992), have suggested that in a single reproductive season cubs can be produced by more than one female within each social group. However, multiple paternity within a litter has not been demonstrated.
Low genetic variability in badgers has been described using allozymes (Evans et al. 1989), multiple and single locus DNA fingerprinting (da Silva et al. 1994; Burke et al. 1996; E. Geffen personal communication), and microsatellites isolated from the Eurasian otter (Lutra lutra, Dallas & Piertney 1998). Random amplified polymorphic DNA markers showed moderate levels of polymorphism (P. Fakler and A. Schreiber, unpublished data). Bijlsma et al. (2000) described seven microsatellite markers isolated from Eurasian badgers that were moderately variable in a sample of 105 badgers from different localities in the Netherlands and Denmark. However, the markers might not be variable enough for paternity testing in a single population or in social badgers, where individuals might be closely related.
The genetic resolution for individual identification and the establishment of familial relationships can be complemented with accurate field records and behavioural observations (Mace et al. 1996). In this study, we use microsatellite DNA technology to detect genetic polymorphism in the badger and combine the results with behavioural and physiological data from Woodroffe (1992) to explore baseline hypotheses of the badger mating system, such as the number of breeding animals. We also examine and discuss the demographic factors and mutation patterns that can be linked to the low genetic variability observed in badger microsatellites.
Materials and methods
We studied a badger population living in Wytham Woods, Oxfordshire, UK, a property of 6 km2 belonging to the University of Oxford. The population density of badgers in this area of woodland, pastures and arable land has been increasing steadily since 1986 (Macdonald et al. 2002) and in 1990 and 1991 when samples for this study were collected, densities were estimated as 22.3 and 27.2 individuals (adults plus cubs) per km2, respectively. Badgers live in setts (identified by two capital letters in this study) distributed across Wytham Woods (Fig. 1). The roads and water bodies around the study area present obstacles and risks for badgers to emigrate from or immigrate to Wytham Woods.
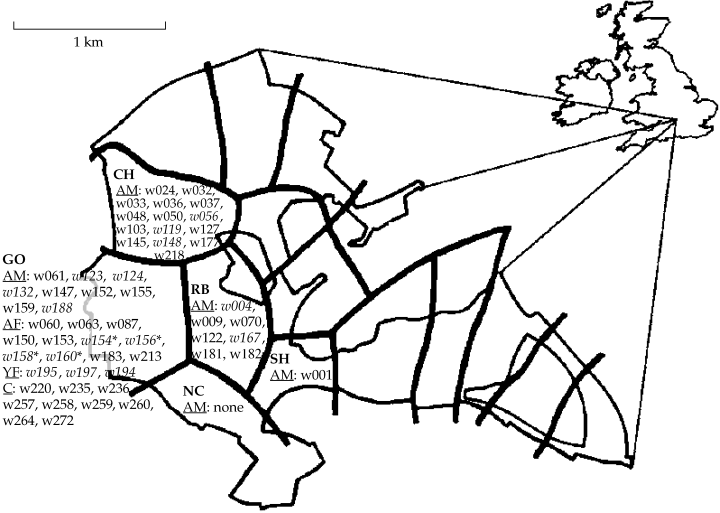
Distribution of Eurasian badger territories (CH, GO, NC, RB and SH) in Wytham Woods in 1991. Thin lines demarcate the limit between wooded areas, where the core of the territories are, and pasture. Thick lines are territory borders as determined by radio-tracking and/or bait-marking. All Wytham badgers have an ID number prefaced with a ‘w’. AM signifies adult males living in the territories during 1990 which are therefore candidate fathers of the offspring born in 1991. AF signifies adult females that could potentially breed; YF signifies young females and C signifies cubs born in GO in 1991. An asterisk indicates females not found to be pregnant in 1991 by ultrasound. Italics represent animals not sampled.
To construct three nonenriched libraries, whole DNA isolated from the muscle of a dead badger (w213) was cut with enzyme AluI or MboI and fragments between 100 and 500 base pairs (bp) were selected. Fragments were ligated into plasmid vector pUC18 and recombinant plasmids were transformed into Escherichia coli XL1-B. Colonies containing microsatellites were detected and selected using probes (GC)10, (GT)10 and (AT)10 (for the first library), (CA)15 (for the second library) and (GAAA)10 and (GATA)10 for two separate hybridizations of the third library. Further details on library construction and selection have been described elsewhere (Domingo-Roura 1998, 2002). Another enriched DNA library was constructed by Genetic Identification Services as described in Kays et al. (2000). Sequences from these libraries for the loci presented in Table 1 have been deposited in the EMBL database under accession numbers AJ309849 and AJ309052–AJ309059.
| Locus | Repeat | Primer sequences (5′ > 3′) | T a (°C) | Product size (bp) | Within population | Across populations | ||
|---|---|---|---|---|---|---|---|---|
| n | No. of alleles | n | No. of alleles | |||||
| Mel1 | (GT)20 | CTGGGGAAAATGGCTAAACC | 60 | 262–274 | 16 | 1 | 105 | 5 |
| AATGCAGGCTTTGCAATTCC | ||||||||
| Mel3 | (GT)13 | CTAAAACCACCACCACAATGC | 62 | 128–134 | 16 | 1 | 105 | 4 |
| GTGTATAGCCTGCGAACAAGG | ||||||||
| Mel7 | (GT)21 | ATTCTTCCTTTTAGCTTTGGCC | 62 | 134–144 | 16 | 1 | 105 | 5 |
| TCTCACAGTGTCAGCAGAAAGG | ||||||||
| Mel10 | (TG3C)2T3CT(TA3)3(AC)14 | TGCAGAAAACGATCAGGTTTC | 58 | 154 | 40 | 2 | 12 | 5 |
| TCATTTTTGGCGTGTACTTCC | ||||||||
| Mel11 | T 3 CT 7 CT 13 | CCTAGTTGGTAAGAGTCAAGAACAC | 59 | 122 | 40 | 1 | 12 | 4 |
| TGGGGAATGACATGCCATAG | ||||||||
| Mel12 | (TTG)2(TG)6TA(TG)20 | TCCGTTTATATTGCTGTGCC | 53 | 153 | 40 | 4 | 11 | 6 |
| CTGCTCAGCCTTCAAGGTAAC | ||||||||
| Mel13 | (AC)6AG(AC)7 | GGAGACATGACCTTGGACATCCTG | 52 | 94 | 40 | 1 | 11 | 4 |
| CTGGGCAGTCATAGTCGTG | ||||||||
| Mel14 | (AC)18 | GACACAAGCAAACTTTCTTCC | 56 | 188 | 40 | 4 | 12 | 7 |
| CATCTGGAAGTAGGCATAATG | ||||||||
| Mel15 | (CT3)14C2T7A3GAT4(AT3)2(GA)7A(AG)4 | ATGGAACCTCCTGGAAAGAG | 61 | 270 | 40 | 4 | 12 | 5 |
| CAGGTCATGATCCCAGAGTC | ||||||||
| Mel16 | T 5(CT3)10CT2C2T2C(CT3)9C(CT2CT3)2 | ACTGTTTCCTAAGACTGCGT | 59 | 324 | 40 | 1* | 12 | 9 |
| CACCTGACTTGTGAGATGAG | ||||||||
| Mel17 | CT 2 CT 3 CT 2 C 4(C2T2)11C3T2C2T3C2T2C T4C2T3C2T2CT2(C3T)2CAT2CT2CT3 | CTTACTGCAAACACTACCA | 52 | 298 | 40 | 1 | 11 | 11 |
| (C2T2)2(C3T)3C2AT6C | ACAGGACAGTCTTCACAGAG | |||||||
| (C2T2)2CTCT2C2T(C3T)2CT3 | ||||||||
| Mel18 | (C2T2)9C(CT)7T3 | ACTGAATACCAACTACGAAC | 52 | 359 | 13 | 1 | ? | ? |
| TTCCAACAGCTAATGACA | ||||||||
- Data for Mel1, Mel3 and Mel7 are from Bijlsma et al. (2000) based on animals from the Netherlands and Denmark with the exception of within population data that was obtained from Wytham badgers. Ta= annealing temperature.
- * No alleles within the Wytham population could be clearly differentiated in 200 × 200 mm 6% polyacrylamide gels but alleles were obtained through sequencing; ? = not available.
Sequences were selected based on the microsatellite structure and a preliminary test of polymorphism of four to 20 unrelated badgers, mostly from separate geographical regions. The loci that showed variability in this initial screening were amplified in a group of 12 badgers with the following geographical origins: Crete, Germany, Norway, Austria, northeast Spain, northcentral Spain, Japan, United Kingdom, Sweden, Finland, Ireland and Mongolia. The loci were also amplified for Wytham males captured in focal sett GO and surrounding setts (CH, NC, RB and SH) in 1990 when matings occurred and for all pregnant and/or lactating females and all offspring captured in GO in 1991. In 1991, 140 of the estimated 163 badgers living in Wytham were captured (see Macdonald & Newman 2002 for details). GO sett was selected because it was intensively observed and trapped during the study period (Woodroffe 1992), securing the sampling of all candidate mothers and cubs. Mel18 was only tested in 13 mothers and offspring from Wytham Woods. In addition to using the markers that we developed, we also amplified and scored 16 individuals including mothers and offspring from GO sett with primers Mel1, Mel3 and Mel7 (Bijlsma et al. 2000). Polymerase chain reaction (PCR) conditions for microsatellite amplification were the same as described by Domingo-Roura (2002) and the annealing temperatures are given in Table 1.
Distribution of microsatellite alleles was visually scored in 200 × 200 × 1.5 mm 6% polyacrylamide gels and stained either with silver nitrate or ethidium bromide. For marker Mel16, different alleles were detected but could not be reliably scored since they were due to nucleotide substitutions within the microsatellites. In this case, PCR products were cleaned with QIAquick PCR Purification Kit (Quiagen) and sequenced using a dRhodamine Terminator Cycle Sequencing Kit (Perkin-Elmer) following the instructions provided by the manufacturer and were run in an ABI Prism 377 DNA Sequencer (Perkin-Elmer). Heterozygotes in the sequences were detected by the presence of two peaks of similar height for two different nucleotides in a single base position. For maternity exclusion, in case of doubt, a conservative reading assuming heterozygosity was scored. Lengths of microsatellite alleles were determined either from allele sequences or using Bio-Rad Gel Doc 2000 and quantity one software (Bio-Rad Technical Service Department 2000).
Maternity by a specific female was excluded if she could not have contributed either allele found in the offspring. For within-population data, allele frequencies of variable loci were computed by the allele count method and their standard deviations were calculated as the square root of the variance of the corresponding binomial proportions. Maximum likelihood haplotype frequencies were estimated following Excoffier & Slatkin (1995). Diversity indices were computed following Nei (1987). Deviations of loci from Hardy–Weinberg equilibrium were conducted with an exact test using a Markov chain algorithm with 10 000 steps (Guo & Thompson 1992). Pairwise linkage disequilibrium was tested following Slatkin & Excoffier (1996) with 10 000 permutations. The power of discrimination (Pod), that is, the probability that two different individuals have different genotypes for a given loci, was calculated following Fisher (1951). The a priori probability of paternity exclusion (Pex) was calculated following Chakraborty & Jin (1993).
Analysis of molecular variance (amova; Excoffier et al. 1992) using Mel10, Mel12, Mel14 and Mel15 alleles was used to test for substructuring in the data set when comparing (i) adult females (n = 7) and cubs (n = 9); (ii) adult males (n = 24) and adult females (n = 7); and (iii) setts CH (n = 12), GO (n = 21), RB (n = 6) and SH (n = 3). Tests were conducted using arlequin version 2.000 (Schneider et al. 2000).
Microsatellites were classified according to Weber (1990) as perfect, imperfect and compound repeat sequences. The χ2 tests were conducted to compare the number of perfect, imperfect and compound dinucleotide-based repeats obtained in this study with published results in other mustelids (Lutra lutra, Dallas & Piertney 1998; Gulo gulo, Davis & Strobeck 1998; Duffy et al. 1998; Walker et al. 2001; Martes americana and Taxidea taxus, Davis & Strobeck 1998; Mustela vison and M. erminea, Fleming et al. 1999) and in other mammalian species (humans, Weber 1990; pigs, Winterøet al. 1992; dogs Ostrander et al. 1993). The mean and standard deviation (SD) of repeat number were compared between our study and others using t-tests. We computed a regression line of maximum repeat length of perfect repeats against expected heterozygosity (HE) using data from other studies on mustelids (Davis & Strobeck 1998; Fleming et al. 1999; Walker et al. 2001). All analyses dealing with microsatellite structure were conducted using spss 9.0 (SPSS Inc.) for Windows.
Results
The three nonenriched libraries combined gave around 4000 clones of which 38 were positives. Twenty-nine microsatellite regions from the nonenriched and four (Mel15, Mel16, Mel17 and Mel18) from the enriched libraries were tested for variability across Europe and/or within Wytham. For locus Mel16, we discovered size homoplasy in Wytham animals. Consequently, we sequenced the PCR products of GO females and offspring to determine alleles and maternity for this locus. Markers Mel1, Mel3 and Mel7, found previously to be polymorphic in badgers from the Netherlands and Denmark, were monomorphic in 16 individuals that we tested from Wytham Woods.
None of the microsatellites that showed size polymorphism in Wytham revealed deviations from Hardy–Weinberg expectations or showed evidence of linkage (α = 0.05). The combination of allele information from Mel10, Mel12, Mel14 and Mel15 in badgers from Wytham Woods (Table 2) showed a reduced number of rare alleles, a mean number of pairwise differences of 2.144, and a mean HE of 0.536. Markers Mel14 and Mel15 showed a bimodal distribution of allele frequencies with no intermediate-sized alleles (Table 2).
| Locus | k | 2n | Allele or haplotype | Frequency (SD) | H O | H E | Pod | Pex |
|---|---|---|---|---|---|---|---|---|
| Mel10 | 2 | 80 | 111 113 | 0.912 (0.032) 0.087 (0.032) | 0.175 | 0.184 | 0.281 | 0.013 |
| Mel12 | 4 | 86 | 116 118 120 122 | 0.395 (0.053) 0.128 (0.036) 0.128 (0.036) 0.349 (0.051) | 0.674 | 0.706 | 0.847 | 0.265 |
| Mel14 | 4 | 86 | 188 190 196 198 | 0.221 (0.045) 0.105 (0.033) 0.570 (0.053) 0.105 (0.033) | 0.581 | 0.612 | 0.795 | 0.199 |
| Mel15 | 4 | 86 | 245 249 257 265 | 0.140 (0.037) 0.151 (0.039) 0.453 (0.054) 0.256 (0.047) | 0.814 | 0.695 | 0.851 | 0.265 |
| Mel16 | 4 | 32 | CTTTA CTTTT TCTTT TTCCT | 0.469 (0.088) 0.062 (0.043) 0.281 (0.079) 0.187 (0.069) | 0.813 | 0.662* | 0.827 | 0.238 |
- * The expected heterozygosity was computed from haplotype frequencies. k, number of allele;
- H O, observed heterozygosity; HE, expected heterozygosity; Pod, power of discrimination; Pex, paternity exclusion.
The overall probability of two random, unrelated individuals having the same genotype for all loci, including Mel16, is 5.79 × 10−4, whereas the overall probability of excluding a candidate parent when the individual is not the parent is 0.674. The probability of an animal being completely homozygous is 0.02. Genetic substructuring in the Wytham dataset gave nonsignificant results for comparisons between adult females and their offspring (P = 0.158) and between adult males and adult females (P = 0.066) but a significant substructuring of the dataset for comparisons among setts (P < 0.001).
Our four libraries yielded six tetranucleotide and 27 dinucleotide repeat loci. Fourteen dinucleotide loci were perfect, 11 were imperfect, and two were part of a compound microsatellite. The number of nonperfect repeat loci is significantly larger than the number obtained from other mustelid species (48 perfect, 10 imperfect, three compound; χ2 = 6.475, P = 0.011) and from other mammalian species (230 perfect, 58 imperfect, 35 compound; χ2 = 4.421, P = 0.035). The average largest repeat number per clone was 11.55 (n = 29, SD = 4.24, range = 6–20), which is significantly different (t = −2.95, P = 0.006) from the values obtained in the other Eurasian badger study (n = 7, mean = 17, SD = 4.32, range = 12–23), and also different (t = −3.69, P < 0.001) from the values from other mustelid species (n = 67, mean = 15.58, SD = 4.87, range = 7–29). The length of the largest repeat number per clone was not different between variable and invariable microsatellite loci in the world sample (t = −0.938, P = 0.357) but was different in the Wytham population (t = −2.72, P = 0.012).
The correlation between expected heterozygosity and the length of the longest perfect repeat in the original clone of published mustelid microsatellites was significant (r = 0.355, P = 0.011) and the regression line followed the equa-tion HE = 0.326 + (0.021 repeat length). Only Mel10 deviates significantly from this regression line.
Using ultrasound to detect pregnancy, we excluded four of 11 reproductive age adult females as candidate mothers in the sett GO (Woodroffe 1992). Four of the offspring (w235, w257, w260 and w272) could be assigned either to female w63 or to female w213 but not to any other candidate mother. Female w63 was ultrasound scanned in January 1991 and she was diagnosed with one foetus. Female w213 could not be ultrasound scanned but had extended teats when she was caught in April 1991, indicating that she was lactating and, thus, a candidate mother. Therefore, these data suggest that w213 was the mother of three cubs. However, the ultrasound examination of w63 was performed at an early stage of pregnancy and additional foetuses could have been missed. In addition, offspring w264 could be assigned to mother w150 since this female was the only one who had a C nucleotide in position 41 bp for primer Mel16. In summary, our data suggest at least three different mothers for the nine offspring captured in GO and outliers during 1991. Finally, we typed males captured in GO and nearby setts during 1990 and 1991 (Table 2), but paternity could not be assigned confidently since some candidate fathers were not sampled (Fig. 1).
Discussion
We found low levels of genetic variability in badgers from Wytham Woods. However, implicit limitations of the study are that results are based on a reduced number of loci and a reduced number of potentially related individuals. Trends in microsatellite evolution and population genetic parameter estimates might be subject to sampling bias and should be taken with caution.
In this study we detected many short microsatellites and a high number of nonperfect repeats, characteristics that are known to be associated with low allelic variation (Weber 1990; Brinkmann et al. 1998). Only two out of seven of the loci described by Bijlsma et al. (2000) were imperfect in the original clones. Some of the results could be explained by a low stringency in the washes during library production that can cause the relative size and number of loci isolated from genomic libraries to vary greatly among experiments and experimenters. It is always risky to compare data from different libraries and across studies but the high frequency of short, complex repeats is not likely to be only due a systematic bias in probing efficiency during library production, since in this study we produced different libraries under different conditions (see Materials and methods section).
Our results, and those of previous studies using different markers (Evans et al. 1989; da Silva et al. 1994; Burke et al. 1996), suggest genome-wide reductions in variability for British badgers. We found lower overall levels of variation and three loci to be monomorphic in the Wytham population that were variable in the Netherlands and Denmark (Table 1, Bijlsma et al. 2000). In addition, Mel14 and Mel15 have alleles of intermediate sizes missing (Table 2) that could have been lost by genetic drift. We also detected significant substructuring by sett in the Wytham population consistent with a strong matrilineal structure as suggested by behavioural data.
The reduced variability we found might be due to two main groups of factors, one dealing with the isolation, structure and evolution of microsatellite regions and the other linked to demographic and selective events. The two groups of factors might have additive effects since, for instance, the variance in repeat number is determined by mutation rate and by effective population size (Estoup & Cornuet 1999). The bias in library production is not likely to be the main factor responsible for this low variability; it is possible that British badgers have indeed suffered reduced population numbers in the recent past. Nevertheless, the inclusion of related animals in the sample as well as the particular badger reproductive and social system might have accentuated this reduction in polymorphism.
The multiple maternity that we found appears to have behavioural implications within setts. Aggression occurring between females or towards the offspring from subordinate females, as is usual in canids (Kruuk 1989; Girman et al. 1997), does not necessarily result in reproductive suppression in the badger.
Our genetic data also support previous behavioural observations suggesting that reproductive output may be skewed, for instance towards older females or females in better nutritional condition (Cheeseman et al. 1987; Woodroffe & Macdonald 1995a). Gestation and lactation are costly in badgers (Cheeseman et al. 1987; Woodroffe & Macdonald 1995b). For females in poor condition the costs of maintaining gestation and the expected cost of lactation may outweigh the potential benefit of breeding, favouring termination of their pregnancies (Clutton-Brock 1991). Additionally, badgers have delayed implantation allowing for more precise timing of development to match environmental conditions (Harrison & Neal 1956; Woodroffe 1995). Because of reduced levels of genetic variation, con-clusive evidence of reproductive skew and detailed mapping of kinship and reproduction in the Wytham Woods population will require an extensive effort to develop additional polymorphic loci to complement detailed demographic and behavioural data.
Acknowledgements
Badger samples from different countries were provided by J. Brabec (University of Innsbruck), H. Brøseth (Norwegian Institute for Nature Research), P. Fakler (Universität Heidelberg), K. Kauhala (Finnish Game and Fisheries Research Institute), A. Laurie (Eastern Steppes Biodiversity Project), M. Miralles (Rectoria Vella), Midori Saeki (WildCRU), Servicio de Ecopatología de Fauna Salvaje (Universitat Autònoma de Barcelona), and P. Sleeman (University College Cork). J. da Silva and C. Newman helped with sample collection and storage. G. Fernández, E. Geffen, A. B. Ibarz, B. Jaén, J. F. López-Giráldez and R. Whiteley conducted part of the experiments. I. Colson, C. Greig, K. Koeffli, J. Maldonado, M. Vallès and N. Ziv offered advice in the laboratory. J. Alcañiz, J. Bertranpetit and D. Goldstein provided logistic support. F. Calafell helped in data analysis and we thank him and four anonymous reviewers for providing useful comments to improve earlier versions of the manuscript. Financial support was provided by Institut d’Estudis Catalans, People's Trust for Endangered Species (UK), Ministerio de Educación y Cultura (Spain) (Ref. PB98–1064), Natural Environment Research Council (UK) (Ref. GT 4/96/242) and the National Science Foundation (US).
References
This research is part of the long-term Wytham badger field project led by David Macdonald and has been a collaboration with the groups led by Terry Burke, Jaume Terradas and Bob Wayne. Rosie Woodroffe, Michael Roy and Josep Marmi are past members of WILDCRU who worked on this project. Xavier Domingo-Roura and Josep Marmi are now at the Universitat Pompeu Fabra and conduct research on the evolution and application of microsatellite technology in mustelids and primates.




