Population structure and biogeography of migratory freshwater fishes (Prochilodus: Characiformes) in major South American rivers
Abstract
Mitochondrial DNA (mtDNA) sequences were used to infer the phylogenetic relationships of Prochilodus species in the Paraná, Amazonas, Orinoco, and Magdalena basins. Sequences of ATPase subunits 6 and 8 (total 840 bp) were obtained for 21 Prochilodus specimens from the four river systems. Using Semaprochilodus as an outgroup, phylogenetic analyses showed that: (i) each river basin contains a monophyletic group of mtDNA lineages; and (ii) the branching order places Magdalena in a basal position with subsequent branching of Orinoco, Amazon and Paraná. The mitochondrial control region was sequenced for 26 P. lineatus (from the Paraná basin) and six other Prochilodus specimens from the Magdalena, Orinoco and Amazon. All 26 control region haplotypes were unique with sequence divergence ranging from 0.3 to 3.6%. The control region phylogeny is well resolved but phylogenetic structure is not associated with geography. For example, mtDNA haplotypes from the upper Paraná (Mogui Guassú) and the upper Bermejo, separated by at least 2600 km, have close genealogical ties. Phylogeographic analyses, including nested clade analysis, suggest high levels of gene flow within this basin.
Introduction
Historical biogeographic analyses of freshwater fishes provide a natural link between the geological and biotic evolution of a region because dispersal of fishes depends on direct connections between river basins, and the history of basin interconnections reflects the underlying geology (Lundberg 1993; Bermingham & Martin 1998). Inferences of historical biogeography are based on phylogenetic patterns shared by multiple codistributed species (reviewed by Humphries & Parenti 1986). Mitochondrial DNA (mtDNA) genealogies have been used extensively to trace processes at the population level and the phylogenetic diversification of taxa in relation to their geographical distribution (‘phylogeography’, Avise et al. 1987; Avise 2000). When mtDNA substitution rates are homogeneous across lineages and time, the chronology of vicariant events and diversification may also be estimated from genetic distance data and compared to known dates of geological events (Bermingham & Avise 1986; Page 1991, 1996). Phylogeographic studies of ubiquitous, widely distributed freshwater species that live in major rivers should reflect patterns of historical geomorphological processes that contributed to present-day physiography and hydrology (Bermingham & Martin 1998). Unfortunately, there is a paucity of such studies on freshwater fish species from the major river systems in South America to allow historical biogeographic analysis. The major drainage systems in South America developed to their present form after several tectonic episodes, starting about 89 million years ago (Ma) and culminating with the final rise of the Andes some 10 Ma. Thus, the evolutionary diversification of the Neotropical aquatic biota has been shaped by multiple occurrences of vicariance as well as drainage system coalescence events (Lundberg et al. 1998). Despite the tremendous impact of Andean tectonics on freshwater fish faunas, no study to date has examined its effects on the distribution of genetic variation and speciation of a monophyletic group of closely related species at a continent-wide scale.
Fishes of the genus Prochilodus are among the most conspicuous, abundant, and widespread freshwater species living in South American rivers flowing to the Atlantic Ocean. They are large migratory detritivores that support important fisheries in many parts of the continent (Welcomme 1979). Over the past 40 years, tagging experiments and observations have provided extensive documentation of the annual movements of large schools of juvenile and adult Prochilodus (e.g. Bonetto & Pignalberi 1964; Godoy 1967; Bayley 1973; Espinach Ros et al. 1990). These fishes typically undertake long-distance upstream migrations before the onset of the rainy season. During the annual cycle, Prochilodus swim hundreds of kilometers along the river, from the feeding grounds in the floodplains, or lower reaches of their range, to the spawning areas close to the headwaters. Prochilodus species have very high fecundity and spawn all the eggs at once in the open waters of the main river channel. Fertilized eggs drift downstream and onto the floodplains where the larvae feed. Their detritivorous habit makes Prochilodus a dominant element in structuring tropical stream community dynamics and ecosystem attributes via sediment processing activities (Bowen 1983; Flecker 1996). In the Paraná basin, P. lineatus may account for up to 50–90% of the total fish biomass in the lower stretches of the river and in the flood plain lagoons (Bonetto 1986; Cordiviola de Yuan 1992).
A recent revisionary study of the genus recognized 13 species (Castro 1990) distributed in all major rivers of South America. Morphological variation among Prochilodus species is limited, and few of the characters considered by Castro (1990) are unambiguously diagnostic at the species level. An underlying assumption for this taxonomic work has been based on the high intrabasin vagility of Prochilodus, such that each river basin is thought to contain a single panmictic population. The resulting taxonomy restricts most species to a single drainage. Notable exceptions are the São Francisco basin from eastern Brazil with three described species, and P. lineatus and P. vimboides reported to be sympatric in the upper Paraná and upper Uruguay rivers (Castro 1990). A more typical pattern is seen in P. nigricans, which is distributed throughout the Amazon basin, probably constituting one of the most widely distributed species of South American freshwater fishes (Castro 1993).
Our work includes samples of P. magdalenae from the Magdalena basin (Colombia), P. mariae from the Orinoco basin (Venezuela), P. nigricans from two distant localities in the Amazon basin (Brasil and Peru), and P. lineatus from the Paraná basin (Fig. 1, Table 1). We present a phylogenetic hypothesis relating these four species in the framework of the geological history of tropical South American rivers (Lundberg et al. 1998). This work is the first attempt to characterize broadscale biogeographic relationships among all major river basins in South America using mtDNA genealogies. A second component of our study is to characterize the population structure of P. lineatus in the Paraná–Paraguay–Uruguay system (henceforth referred to as Paraná). In addition to traditional phylogeographic analysis (Avise et al. 1987; Avise 2000), we apply ‘nested clade analysis’ (Templeton et al. 1995) as a tool to distinguish between recurrent gene flow and historical events shaping population structure, such as colonization and range expansion. The Paraná is the second largest basin in South America and drains about four million square kilometers of Argentina, Bolivia, Brazil, Paraguay, and Uruguay. Specimens of P. lineatus collected throughout this area (Fig. 1) provide the opportunity to test the idea that these highly migratory fish may indeed form a single panmictic population across such a huge river system.
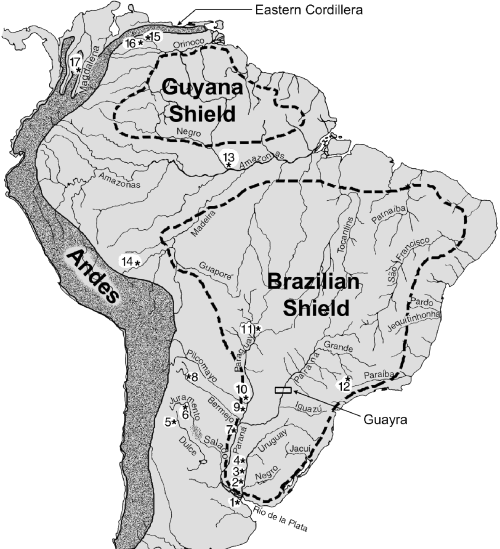
Sampling locations (numbered asterisks 1–17) and major geographical and geological features affecting the distribution of Prochilodus species. Also see Table 1.
| Species | Locality number | Locality | Control region* | ATPase (Haplotypes) | Catalogue number |
|---|---|---|---|---|---|
| P. lineatus | 1 | Buenos Aires, Río de la Plata, ARG | 3 | 1 (H1) | USNM 361375 |
| P. lineatus | 2 | Gualeguaychu, Río Uruguay, ARG | 3 | 1 (H8) | |
| P. lineatus | 3 | Salto Grande, Río Uruguay, ARG | 4 | 1 (H10) | USNM 361378 |
| P. lineatus | 4 | Monte Caseros, Río Uruguay, ARG | 2 | — | |
| P. lineatus | 5 | Termas, Río Dulce, ARG | 1 | — | USNM 361245 |
| P. lineatus | 6 | El Tunal, Río Juramento, ARG | 1 | — | USNM 361250 |
| P. lineatus | 7 | Bella Vista, Río Parana, ARG | 1 | 4 (H1, H2, H3, H6) | |
| P. lineatus | 8 | Embarcacion, Río Bermejo, ARG | 4 | 1 (H1) | USNM 361249 |
| P. lineatus | 9 | Riacho Mercedes, Río Paraguay, ARG | — | 2 (H1, H9) | |
| P. lineatus | 10 | Asuncion, Río Paraguay, PAR | 2 | 1 (H5) | |
| P. lineatus | 11 | Corumba, Río Paraguay, BRA | 3 | 2 (H4, H7) | USNM 361376 |
| P. lineatus | 12 | Represa Beija-Flor, Río Mogui Guassú, BRA | 2 | — | |
| P. nigricans | 13 | Manaus, Río Amazonas, BRA | 1 | 1 (H11) | |
| P. nigricans | 14 | Manu, Río Madeira, PER | — | 2 (H12) | |
| P. mariae | 15 | Portuguesa, Río Apure, VEN | 1 | 2 | |
| P. mariae | 16 | Río Las Marias, VEN | 2 | — | |
| P. magdalenae | 17 | Guataperi, Río Magdalena, COL | 2 | 3 | |
| Semaprochilodus | n/a | Manaus, Río Amazonas, BRA | — | 2 |
- * Each control region sequence constituted a unique haplotype.
Methods
To study phylogeny and assess the biogeographic pattern of Prochilodus in major South American rivers, complete nucleotide sequences (840 bp) of the slightly overlapping mitochondrial genes for ATP synthase subunit six (ATPase6) and eight (ATPase8) were obtained from 21 specimens. These consisted of 11 P. lineatus from the Paraná basin, two P. nigricans from the Amazon, two P. mariae from the Orinoco, and three P. magdalenae from the Magdalena drainage (Table 1). Two Semaprochilodus specimens were sequenced to provide an outgroup. To characterize finer level population structure within the Paraná basin, the hypervariable mitochondrial control region (that includes the D-loop) was sequenced from 26 P. lineatus sampled from 12 widespread Paraná localities. Three P. mariae, two P. magdalenae, and one P. nigricans were sequenced to serve as outgroup taxa. These sequences represent the complete mtDNA control region (approximately 1100 bp) along with the flanking tRNA genes — about 20 bp of tRNA Thr (3′ half), the complete 72 bp of tRNA Pro, about 65 bp tRNA Phe (almost complete).
Genomic DNA was isolated from ethanol-preserved muscle tissue by standard proteinase K, phenol–chloroform extraction (Sambrook et al. 1989). The mitochondrial ATPase6 and 8 genes were amplified using the primers ATP 8.2_L8331 (5′ AAAGCRTYRGCCTTTTAAGC) and CO3.2_H9236 (5′ GTTAGTGGTCAKGGGCTTGGRTC), and were sequenced using the aforementioned primers plus the internal primer ATP8.3_H9407 (5′ AAAGTTCCTGTGGTGTGCGGGGG) according to the general methods described in Lovette et al. (1998). The mitochondrial control region was amplified by polymerase chain reaction (PCR) in 50 µL reactions containing 10 µL dNTPs (1 m m), 5 µL reaction buffer (200 m m Tris-HCl pH 8.4, 500 m m KCl), 2 µL MgCl2 (50 m m), 2 µL of each primer (10 µm), 0.5 µL (2.5 U) of Taq DNA polymerase (Gibco BRL), 2 µL of template DNA (100 ng/µL) and 26.5 µL of H2O. PCR conditions were as follows: 94 °C (3 min), 10 cycles of 94 °C (1 min), 53 °C (1 min), 72 °C (1 min), 10 cycles of 94 °C (1 min), 51 °C (30 s), 72 °C (1 min), 10 cycles of 94 °C (1 min), 50 °C (30 s), 72 °C (1 min), followed by 72 °C (2 min). The following primers were used for PCR and sequencing: F-TTF: 5′ GCCTAAGAGCATCGGTCTTGTAA and F-12R: 5′ GTCAGGACCATGCCTTTGTG. Additional internal primers used for sequencing were PDF2: 5′ TCTATGCAAAGATCCAACTA, PDR2: 5′ GTGGTTATTTAAGCAATGTC, and PDR2-2: 5′ GAGAGTGTATGCACCTGAT. Samples were sequenced using the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems Inc.) on an ABI 310 automated DNA sequencer following manufacturer’s instructions. All templates were sequenced completely in both directions. The nucleotide sequence data have been deposited in GenBank (accession numbers: AF281827–AF281849 for ATPase; AF282733–AF282764 for control region).
Phylogenetic methods
Control region sequences were aligned using ClustalW (Gibson et al. 1996). Settings for clustalw were opening gap cost = 20, extending gap cost = 5 for both pairwise and multiple alignments. ATPase sequences were aligned by eye and checked for accuracy using the translated protein sequences. Phylogenetic analyses were conducted separately for each data set using paup* version 4.0b4 (Swofford 2000). We also analysed the combined data for the 10 individuals sequenced for both control region and ATPase6,8, and used the partition homogeneity test of Farris et al. (1994) to gauge incongruence between data sets. Maximum parsimony (MP) analyses used heuristic searches starting with stepwise addition trees and replicated 100 times, with each replicate starting with random input order of sequences. Branch swapping was performed by the tree-bisection-reconnection (TBR) method using default parameters. Bootstrap analysis was used to gauge support of the resulting topology and was based on 1000 replicates of heuristic search with starting trees obtained by stepwise addition and TBR branch swapping. Modeltest 3.0 (Posada & Crandall 1998) was used to determine the optimal model of nucleotide evolution for each data set. The parameter values estimated by Modeltest 3.0 were used to perform maximum likelihood (ML) and minimum evolution (ME; Rzhetsky & Nei 1992) heuristic searches. Bootstrap analyses also were performed using minimum evolution as the optimality criterion, with the same settings as the MP bootstrap analyses. As an additional test for geographical structure within the Paraná basin, a constrained MP search was done using the control region data, forcing haplotypes from the same geographical locality to lie within a clade. This tree was compared with the unconstrained MP tree using Templeton’ (1983) Wilcoxon signed-ranks test and Kishino–Hasegawa’s parametric test (Kishino & Hasegawa 1989).
Nested clade analysis
Nested clade analysis uses an intraspecific cladogram estimation procedure designed to circumvent difficulties common to many applications of intraspecific phlyogenetics. Some of these difficulties include low variation among haplotypes and the treatment of ancestral types. This approach allows efficient use of geographical information in a genealogical hypothesis-testing framework (Crandall & Templeton 1996). Given that ATPase haplotype diversity was low, and that phylogenetic relationships among haplotypes from the Paraná samples remained largely unresolved, nested clade analysis was considered suitable to analyse further the ATPase data. The cladogram estimation procedure and nesting rules described by Templeton et al. (1992, 1995), and extended for sequence data by Crandall (1996), were used to construct the nested cladogram set. The limits of parsimony for the data set were calculated and a network subsequently constructed with connections having probabilities greater than 0.95, using the program tcs (version 1.0) by M. Clement, D. Posada, and K. A. Crandall (http://bioag.byu.edu/zoology/crandalllab). The resulting genealogical links were checked manually against a distance matrix and errors were corrected by hand. Nested clade analysis uses this haplotype network to define a series of nested branches or clades. This nesting structure, together with information on geographical distribution of the haplotypes, is used to estimate two geographical measures for each clade, the clade distance (Dc) and the nested clade distance (Dn). Dc is a measure of the geographical extent of a given clade, while Dn is a measure of the average geographical distance of individuals in a clade from those in the next higher-level nesting clade within which it is contained (Templeton 1998). Geographic analyses were performed using GeoDis 2.0 (Posada et al. 2000). Distances among sampling locations were estimated on a map following the course of rivers (Table 2). The inference key provided by Templeton (1998) was used to interpret the outcome of the geographical association analysis.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | |||||||||||||
| 2 | 146 | — | ||||||||||||
| 3 | 390 | 244 | — | |||||||||||
| 4 | 430 | 284 | 40 | — | ||||||||||
| 5 | 1176 | 1322 | 1566 | 1606 | — | |||||||||
| 6 | 1360 | 1506 | 1750 | 1790 | 816 | — | ||||||||
| 7 | 820 | 966 | 1210 | 1250 | 1412 | 1596 | — | |||||||
| 8 | 1816 | 1962 | 2206 | 2246 | 2408 | 2592 | 996 | — | ||||||
| 9 | 1306 | 1452 | 1696 | 1736 | 1898 | 2082 | 486 | 1026 | — | |||||
| 10 | 1306 | 1452 | 1696 | 1736 | 1898 | 2082 | 486 | 1026 | 50 | — | ||||
| 11 | 2284 | 2430 | 2674 | 2714 | 2876 | 3060 | 1464 | 2004 | 1028 | 978 | — | |||
| 12 | 2764 | 2910 | 3154 | 3194 | 3356 | 3540 | 1944 | 2664 | 2154 | 2154 | 3132 | — | ||
| 13 | 5212 | 5358 | 5602 | 5642 | 5804 | 5988 | 4392 | 4932 | 3956 | 3906 | 2928 | 6060 | — | |
| 14 | 4564 | 4710 | 4954 | 4994 | 5156 | 5340 | 3744 | 4284 | 3308 | 3258 | 2280 | 5412 | 2376 | — |
Results
ATPase sequence variation
ATPase sequence divergence (uncorrected) ranged from 0 to 5% among mtDNA lineages within Prochilodus, and from 12.5 to 16% between Prochilodus and the Semaprochilodus lineages used to root the tree. Among the 13 samples of P. lineatus from the Paraná basin, maximum divergence was 1.3% and four sequences were identical (haplotype H1, Table 1). The identical haplotypes were obtained from geographically distant localities, separated by a maximum distance of 1816 km. The two sequences of P. nigricans from the Madeira river (Manu) also were identical and differed by 0.6% from another P. nigricans haplotype sampled from the Amazonas river at Manaus (Table 1). Haplotypes representing P. magdalenae (Magdalena basin in Colombia) diverged from all other Prochilodus mtDNA haplotypes by at least 4%. The genetic distances between fish in the Paraná, Amazonas and Orinoco basins were lower, ranging from 0.8% to 2.5%.
Among 840 bp of ATPase6 and 8 genes sequenced for 23 individuals, 195 sites were variable, and 111 were parsimony informative. The MP analysis (1000 replications of heuristic searches) resulted in 131 equally parsimonious trees, which differed only in the placement of the various haplotypes of P. lineatus. In all cases, haplotypes representing different river basins (and different nominal species) formed monophyletic groups. mtDNA haplotypes from the Magdalena basin were placed in a basal position with subsequent branching of Orinoco, Amazon, and Paraná sequences. The overall branching order of the strict consensus of all MP trees was completely congruent with the ML tree shown in Fig. 2. The ML tree was estimated using the Hasegawa–Kishino–Yano (HKY85 + gamma) model of nucleotide substitution with among site rate variation (Hasegawa et al. 1985; Yang 1993). This was the simplest model that fit the Prochilodus data using the log-likelihood significance criteria imposed by Modeltest (Posada & Crandall 1998). The following parameters for this model were obtained: transitions/transversions = 10.985, base frequencies A = 0.282, C = 0.315, G = 0.118, T = 0.285, and gamma shape parameter = 0.266.
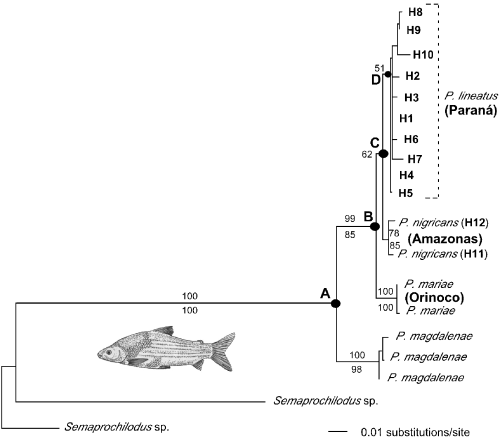
Maximum likelihood tree for ATPase haplotypes (haplotype information in Table 1). The HKY85 + gamma model was used, with the following parameters: transitions/transversions = 10.985, base frequencies A = 0.282, C = 0.315, G = 0.118, T = 0.285, and gamma shape parameter = 0.266. Bootstrap values (>50% only) from maximum parsimony and minimum evolution are shown above and below the branches, respectively. Nodes labelled A–D correspond to separation of lineages among river basins (see text).
ATPase nested clade analysis
Because very little resolution was obtained with the above methods, variation among the 10 unique P. lineatus ATPase haplotypes (H1–H10) and the two P. nigricans haplotypes (H11 and H12) was analysed further with nested clade analysis. The limit for parsimonious connections for this data set (0.95 level) was 12 mutational steps, and the resulting network contained no nonparsimonious connections or any loops of ambiguity. The nested clade structure is shown in Fig. 3. Terminal haplotypes that differ by one substitution were grouped in one-step clades and numbered 1–1 through 1–17 in Fig. 3. Among all one-step clades, only groups 1–1 and 1–9 contained both geographical and genetic variation. Clade 1–3 with haplotype H1 only contained geographical variation because this haplotype was found in four different localities (Table 1). Haplotype H8 (lower Uruguay river) was placed at the root of the P. lineatus subtree because it provided the most parsimonious connection between the Paraná and Amazon haplotypes. Interestingly, control region haplotypes obtained from the lower Uruguay and Río de la Plata specimens were also the most basal branches in the P. lineatus control region genealogy (see below, Fig. 4). Permutation analyses based on 10 000 resamples estimated nonsignificant values of Dc and Dn for clades 2–3, 2–6 and 3–2. Hence, for these clades, the null hypothesis of no association between genealogy and geography could not be rejected. Among all other nested clades, three contiguous range expansion events were inferred from the analysis (Table 3). Significant among these was clade 2–1, which contained haplotypes from several widespread localities in the Paraná system, ranging from the headwaters of the Paraguay river in Corumba (haplotype H4) to the Río de la Plata (H1), covering over 2000 km of river distance. Clearly, no population subdivision among fishes from the Paraná basin was detected by the nested clade method of analysis.
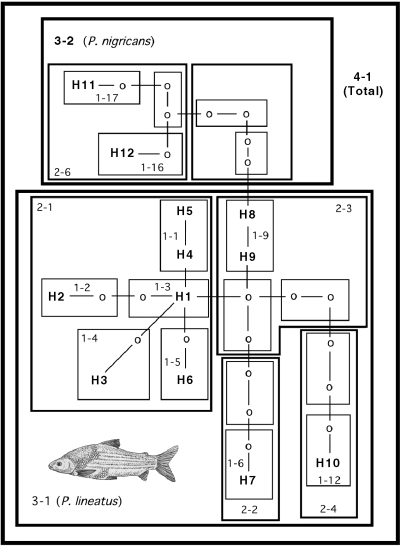
Nested clade diagram for Prochilodus ATPase haplotypes. Haplotype designations follow Table 1. Haplotype H1 occurred in four different locations within the Paraná basin. Missing intermediate haplotypes are indicated by ‘o.’
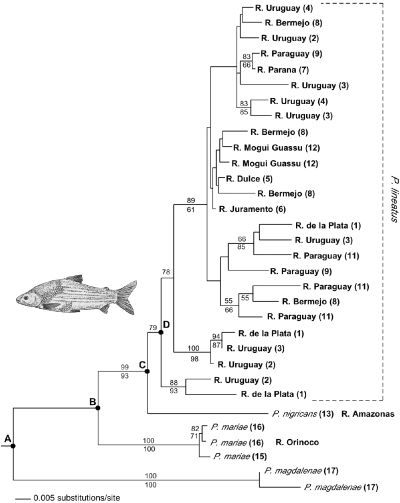
Maximum likelihood tree for Prochilodus control region sequences. The HKY85 + I + gamma model was used with the following parameters: transitions/transversions = 6.522, base frequencies A = 0.333, C = 0.211, G = 0.137, T = 0.319, proportion of invariant sites = 0.6277, and gamma distribution shape parameter = 0.5985. Bootstrap values (> 50% only) from maximum parsimony and minimum evolution are shown above and below the branches, respectively. Locality numbers shown in parentheses follow Table 1 and Fig. 1. Nodes labelled A–D correspond to separation of lineages among river basins (see text).
| Clade | Inference chain | Inferred pattern |
|---|---|---|
| 1–1 | 1–2-3–5-15–16–18-No | Not able to distinguish between range expansion, fragmentation, and isolation by distance |
| 1–9 | 1–2-11–12-No | Contiguous range expansion |
| 2–1 | 1–2-11–12-No | Contiguous range expansion |
| 3–1 | 1–2-11–17-No | Inconclusive outcome |
| Total (4–1) | 1–2-11–12-No | Contiguous range expansion |
| 2–3, 2–6, 3–2 | Fail to reject H0 |
- H0, null hypothesis.
Control region phylogeny
As expected, sequence divergence among control region haplotypes was much higher than for the ATPase genes (Meyer 1993). All sequenced individuals had unique haplotypes, with sequence divergence ranging from 0.3 to 3.6% among P. lineatus, the maximum divergence being nearly three times as large as the maximum divergence for conspecific ATPase sequences. The outgroup lineages for the control region analysis consisted of two P. magdalenae, three P. mariae and one P. nigricans. Sequence divergence between P. lineatus and the outgroup taxa ranged from 3.6 to 8.3%.
The alignment of the control region sequences consisted of 1275 positions, with 223 variable sites. Of these, 152 were parsimony informative. MP analysis yielded 69 equally parsimonious trees. Topological differences among equally parsimonious trees were restricted to some of the less divergent haplotypes within the P. lineatus clade. Control region sequences from each river basin formed a monophyletic group, and the branching order among control region haplotypes representing different river basins matched the order observed across the ATPase-based tree (see 2, 4). Only 10 individuals (nine P. lineatus and one P. nigricans) were sequenced for both ATPase and control region. A combined analysis of these sequences showed no conflict in phylogenetic signal between these markers (nonsignificant partition homogeneity test, Farris et al. 1994). A maximum parsimony search also was performed enforcing a ‘geographical constraint’, under which haplotypes from each locality were forced to group together. This search yielded 20 equally parsimonious trees 461 steps long, about 19% longer than the unconstrained tree (389 steps). This difference is highly significant (Kishino–Hasegawa test, Templeton test, both P < 0.0001), suggesting no significant association between Paraná control region genealogy and geography.
The HKY model with invariant sites and among site rate variation (Hasegawa et al. 1985; Yang 1993; Gu et al. 1995) was used for ML and ME analyses. The ML tree (Fig. 4) showed well-resolved relationships within P. lineatus and overall topology congruent with the strict consensus of all MP trees. Major differences occurred in the placement of some derived lineages within P. lineatus. Like in the MP trees, the ML tree indicated that the basal control region lineages were sampled from P. lineatus collected in the lower and middle Uruguay river and Río de la Plata (localities 1, 2 and 3). However, haplotypes from these localities were also represented among more derived lineages. There was no strong association between the phylogenetic relationships among haplotypes and their geographical distribution. For example, closely related haplotypes collected in the upper Bermejo and in the Mogui Guassú (localities 8 and 12, respectively), were separated by a distance of over 2600 km. Nested clade analysis was not performed on the control region data set because the high level of genetic divergence resulted in nonparsimonious connections for most haplotypes and no nested cladogram set could be obtained (Templeton et al. 1992). At least our qualitative assessment of phylogeographic structure of control region sequences does not seem to counter conclusions drawn from nested clade analysis of the ATPase sequences.
Discussion
Prochilodus lineatus phylogeographic pattern in the Paraná
High levels of mtDNA variation were observed among control region haplotypes sampled throughout the Paraná basin. The intraspecific mtDNA phylogeny was moderately well resolved (Fig. 4), but no significant association between genealogy and geographical location was found. The same finding was supported by nested cladistic analysis of ATPase sequences, albeit with a smaller sample size. Biogeographic studies have consistently identified the Upper Paraná region north of the Guayra rapids (Fig. 1) as a region of endemism distinct from the rest of the basin (e.g. Ringuelet 1975; Géry 1984). However, Prochilodus lineatus sampled from the Upper Paraná (Mogui Guassú river, Fig. 1 locality 12) had control region haplotypes closely related to haplotypes found some 2600 km away, in the Bermejo, Juramento and Dulce rivers, western tributaries of the lower Paraná basin (1, 4, localities 5, 6 and 8). This finding is consistent with the revised taxonomy proposed by Castro (1990) in which P. scrofa, traditionally assigned to the Upper Paraná only, and P. lineatus and P. platensis from the lower Paraná and Paraguay basins were treated as synonyms. Although our samples were limited to a single locality in the Upper Paraná (Locality 12), a recent allozyme survey of P. lineatus revealed high levels of genetic variation but lack of population subdivision among 160 Upper Paraná samples (Revaldaves et al. 1997). Prochilodus are known for their great swimming capacity and their ability to migrate upstream and even overcoming rapids and falls (Géry 1977). The migratory habits of Prochilodus may explain the lack of concordance between the observed pattern of mtDNA variation and the reported areas of endemism based on distributions of many species with limited vagility. However, it cannot be determined whether the mtDNA phylogeographic pattern is a consequence of long-term historical gene flow across the former Guayra rapids that separated the Upper and Lower Paraná regions, or the effect of the recently constructed fish ladder at the Itaipú hydroelectric complex (Borghetti et al. 1994). Distributional limits of many fishes restricted to the Upper or Lower Paraná have been blurred after the Itaipú reservoir covered the Guayra falls, a former natural barrier between these two areas of endemism (Bonetto et al. 1989).
A recent review (Avise 2000) reported that among widely distributed freshwater fish species, studied over a large portion of their range, deep phylogenetic divisions in mtDNA usually occur among geographical areas (‘Category I’sensuAvise et al. 1987). Over half of the species surveyed displayed a Category I phylogeographic pattern. But cases of deeply separated mtDNA lineages with sympatric distributions were uncommon when extensive sampling throughout the species’ range had been conducted (category II: ‘deep gene tree, major lineages broadly sympatric’). Many other species exhibited shallower phylogenetic structures (Categories III to V) when characterized at more restricted spatial scales. mtDNA studies of freshwater fishes that resulted in shallow phylogenetic trees lacking spatial structure have often involved surveys conducted over a relatively small portion of the total range of the species. This was not the case in our study, where a vast geographical sample throughout the Paraná basin failed to show any deep division in the ATPase genealogy (maximum of 1.3% sequence divergence) and any geographical orientation. In fact, nested clade analysis detected evidence for contiguous range expansion within clades 1–9 and 2–1 (Fig. 3), with ‘star phylogeny’ patterns reminiscent of category IV (Avise 2000). This was not due to lack of resolving power of the ATPase marker. Longer and hyper-variable control region sequences characterized for more individuals provide increased power to detect statistically significant lineage structure. Yet, the control region data were clearly consistent with high gene flow or range expansion events maintaining the pattern of variation within this basin in the face of a three-fold increase in the level of genetic variation (0.3–3.6% sequence divergence). Divergent control region haplotypes were found among sympatric P. lineatus sampled from the lower Uruguay river and the Río de la Plata region (Fig. 4). The vast area drained by the Paraná basin and the high vagility of Prochilodus within the basin may have precluded separation of subpopulations for long evolutionary periods. The amount of mtDNA polymorphism is related to the effective size of female populations over evolutionary time (Nei 1987; Avise et al. 1988). Large population sizes of Prochilodus throughout its range simply may account for the high level of mtDNA polymorphism observed.
Although dense sampling for the present study was restricted to the Paraná and P. lineatus, the pattern of genetic continuity throughout a drainage may be general for Prochilodus. Our samples of P. nigricans, an extremely widespread Amazonian species, were drawn from two sites separated by 2376 kilometers, yet also carried ATPase lineages that were only 0.6% different from one another (Fig. 2). mtDNA differences between P. mariae collected from two different tributaries of the Orinoco river were also less than 1%. However, the prevailing taxonomy of Prochilodus indicates some notable exceptions to the extensive geographical continuity of evolutionary lineages documented by our mtDNA results. P. britskii recently has been described as a new species with a restricted distribution in a tributary of the Tapajós, within the Amazonas system (Castro 1993), and three species of Prochilodus are currently assigned to the relatively smaller São Francisco basin (Castro 1990). Nonetheless, the implication that large, abundant, and highly migratory fishes may effectively form a single panmictic population throughout vast drainage areas makes Prochilodus species outstanding biological indicators to trace the history of interconnection across the major drainages of South America.
South American biogeography
This is the first molecular phylogenetic analysis of widespread freshwater fish species distributed over a large spatial scale, involving the major river systems of South America. The mtDNA-based hypotheses of Prochilodus species relationships (2, 4) are consistent with geological evidence documenting the rise of the Eastern Cordillera of Colombia some 10 Ma (Lundberg et al. 1998) as a primary vicariant event, isolating fish populations in the Magdalena system. The geological history of separation and interconnection between the Orinoco and Amazonas, and between the Amazonas and Paraná systems is more complex. Although the Amazonas and the Orinoco drainages remain connected through the Casiquiare river, Prochilodus sampled from these drainages have been assigned to different species (Castro 1990) and also form separate mtDNA clades (2, 4). Less clear is the relationship between the Amazonas and the Paraná, presently separated by a divide in the Pantanal region, between the Andes and the Brazilian shield. At least four headwater-capture events by these river systems have been documented for the past 70 Myr (Lundberg et al. 1998), the most recent during the last 10 Myr. P. nigricans and P. lineatus, assigned to the Amazonas and the Paraná, respectively (Castro 1990), also are seen to form separate mtDNA clades with a sister-group relationship (2-4). Thus, the branching order of the Prochilodus species studied here is consistent with the following model of South American river relationships: {Magdalena [Orinoco (Amazonas, Paraná)]}.
Under the assumption of a molecular clock, mtDNA data can provide information about branching time in addition to branching order. In turn, the chronology of diversification of the separation of mtDNA lineages can be compared to known dates for geological events. In order to test the null hypothesis of a molecular clock (rate constancy across lineages of mtDNA haplotypes), a log likelihood ratio test (Huelsenbeck & Crandall 1997) has been performed. This test showed no significant differences between the likelihoods obtained by enforcing the molecular clock on the topologies shown in 2, 4 and the likelihoods of these trees without the clock enforced. Therefore, node-to-tip distances for the main splitting events in the phylogeny that separate haplotypes from different rivers (2, 4, nodes labelled A–C) can be calculated using ML. These tree-based distances (Table 4) can in turn be translated to rough time-estimates, given an independent dating based on geological evidence. Using the separation of the Amazonas/Orinoco from the Magdalena basin ~10 Ma (Lundberg et al. 1998) as an external calibration point, we obtained values ranging from 3.9 to 5.2 Myr for the separation of Orinoco vs. Amazonas/Paraná, and 2.3–4.1 Myr for the split between Amazonas and Paraná lineages (Table 4). The estimated time of coalescence among Paraná lineages is 1–3.3 Myr (based on the node-to-tip distance for node D).
| Divergence (node in tree) | ATPase (Ma) | Control region (Ma) |
|---|---|---|
| Magdalena vs. others (A) | 5.37 (10) | 16.68 (10) |
| Orinoco vs. Amazon + Paraná (B) | 2.09 (3.9) | 8.72 (5.2) |
| Amazon vs. Paraná (C) | 1.22 (2.3) | 6.90 (4.1) |
| Among Paraná lineages (D) | 0.56 (1.0) | 5.45 (3.3) |
Some caveats apply to our estimates of divergence times. The rates of sequence divergence obtained inferred using the Magdalena vicariance event (10 Ma), are 0.54% per Myr for the ATPase genes and 1.67% per Myr for the D-loop. This difference is consistent with an expected three- to five-fold increase in the rate of substitution in the noncoding control region as compared to protein coding sequences in the mitochondrial genome of other animals (e.g. Meyer 1993; Avise 2000; references therein). However, this substitution rate is less than one half of the 1.3% per Myr divergence rate reported for fish ATPase genes by Bermingham et al. (1997), and less than conventional estimates of substitution rates for protein coding genes in the mitochondria of vertebrates (Brown et al. 1979; Martin et al. 1992). Evaluating reported cases of molecular divergence rates implied by fossil evidence, Lundberg (1998, pp. 53–55) estimated similarly low divergence rates (0.21–0.26% per Myr) for mitochondrial ribosomal genes in other characiform fishes (serrasalmins). These observations raise the intriguing possibility that characiform fishes may have slower rates of mtDNA substitution. Applying the 1.3% per Myr ‘fish’ divergence rate for ATPase (Bermingham et al. 1997), the estimated date of divergence between lineages of Prochilodus in the Magdalena vs. Amazonas/Orinoco is 4.1 Ma. This date does not seem compatible with geological evidence suggesting that the final rise of the Eastern Cordillera and the establishment of the west-to-east flow of the Amazonas and Orinoco were completed by 8 Ma (Lundberg et al. 1998). Whether the rate of nucleotide substitution in the mtDNA ATPase6,8 genes of Prochilodus is slow, or the separation of Magdalena Prochilodus postdates the rise of the Eastern Cordillera of Colombia cannot be determined from the data in hand. Future work comparing sequence divergences among other putative vicariant pairs across this divide could test the possibility of post-Miocence biotic leakage between Magdalena and Orinoco/Amazonas basins, suggested by the low sequence divergence reported here for Prochilodus.
Several biogeographic studies have been published for widespread Neotropical freshwater fish taxa. Hrbek & Larson (1999) studied the phylogeny of the Rivulidae (Cyprinodontiformes) and its relation to geographical distribution. They found the basal taxa in South America associated with the older geological formations (Brazilian and Guyana Shields). Rivulids are small fishes that occupy a diversity of ecological habitats, including ephemeral bodies of water, facilitated by their ability to undergo developmental diapause. Therefore, the value of these taxa as biogeographic indicators for the major river systems is somewhat complicated by this unusual life history pattern. Vari (1988, 1989) studied phylogenetic patterns for the Curimatidae, a large and diverse family of characiform fishes from lowland habitats in South America. Curimatidae is the sister group of Prochilodontidae (Vari 1983). Within the Curimatidae, a phylogeny of the species of the genus Potamorhina shows a basal split between fish of the Maracaibo basin (to the west of the Eastern Cordillera) and species distributed East of the Andes, in the Orinoco, Amazonas, and Paraná basins (Vari 1988). The rise of the Andes likely contributed the basal vicariant event in this genus, similarly to what we report for Prochilodus. However, for the genus Curimata, most speciation events clearly predated the uplift of the Andes (Vari 1989). In general, for most Neotropical freshwater fish groups, Late Miocene through Holocene earth history events (10–2 Ma) have played little or no role in generating much diversity at the family or genus level (Lundberg 1998). Thus, species-level diversification patterns for groups such as Prochilodus and other similarly widespread taxa with comparable life history patterns and ecological characteristics would most likely reflect the recent biogeographic history of major river drainages in South America and contribute to form our picture of landscape evolution in the lowland neotropics.
Acknowledgements
Many colleagues contributed valuable specimens for the study or collaborated during field collections. Our thanks go to A. Espinach Ros, S. Sverlij, R. Delfino, G. Seigneur, A. Fortuny, M. Di Bitetti, A. Balabusic and P. Cichero (Administración de Parques Nacionales, Argentina), A. Catella and A. Moraes (CPAP/EMBRAPA, Corumbá), J. Porto (INPA, Manaus), F. Galeano Vera (Dept. de Acuicultura, Paraguay), J. Dergam, A. Flecker, G. Zuleta, E. Ortí, R. Ortí, C. Chiaramonte, and M. Halpern. Funding for this project was provided by a National Science Foundation grant (INT9117104) to M.A. Bell, and funds from the University of Nebraska-Lincoln, the Smithsonian Institutions Scholarly Studies Program, and the Smithsonian Tropical Research Institute. We thank Gustavo Ybazeta and Nimiadina Gomez (STRI), and A. Meyer (Stony Brook) for their excellent assistance in the laboratory. Insightful comments on earlier versions of this manuscript were provided by J. Lundberg, R. Vari, and two anonymous reviewers.
References
This work was carried out in the laboratories of Guillermo Ortí at the University of Nebraska and of Eldredge Bermingham at STRI. G. Ortí and E. Bermingham have a common interest in studying the evolution and diversification of Neotropical freshwater fishes using molecular markers. Although they have shared collection trips in South America this is their first joint written contribution. More information about research programmes at the laboratories of G. Ortí and E. Bermingham, as well as technical information and aligned data sets may be found at http://go-lab.unl.edu and http://nmg.si.edu/bermlab.htm. This work is part of Arjun Sivasundar’s MS thesis at UNL. Arjun is currently a PhD student at Rutgers University.




