Fine-mapping of the intestinal receptor locus for enterotoxigenic Escherichia coli F4ac on porcine chromosome 13
Summary
The aim of this study was to refine the localization of the receptor locus for fimbriae F4ac. Small intestinal enterocyte preparations from 187 pigs were phenotyped by an in vitro adhesion test using two strains of Escherichia coli representing the variants F4ab and F4ac. The three-generation pedigree comprised eight founders, 18 F1 and 174 F2 animals, for a total of 200 pigs available for the linkage analysis. Results of the adhesion tests on 171 F2 pigs slaughtered at 8 weeks of age show that 23.5% of the pigs were adhesive for F4ab and non-adhesive for F4ac (phenotype F4abR+/F4acR–; R means receptor). Pigs of this phenotype were characterized by a weak adhesion receptor for F4ab. No pigs were found expressing only F4acR and lacking F4abR. Receptors for F4ab and F4ac (F4abR+/F4acR+) were expressed by 54.5% of the pigs. Animals of this phenotype strongly bound both F4ab and F4ac E. coli. In the segregation study, the serum transferrin (TF) gene and 10 microsatellites on chromosome 13 were linked with F4acR (recombination fractions (θ) between 0.00 and 0.11 and lod score values (Z) between 11.4 and 40.4). The 11-point analysis indicates the F4acR locus was located in the interval S0068-Sw1030 close to S0075 and Sw225, with recombination fractions (θ) of 0.05 between F4acR and S0068, 0.04 with Sw1030, and 0.00 with S0075 and Sw225.
The lack of pigs displaying the F4abR–/F4acR+ phenotype and the presence of two phenotypes for F4abR (a strong receptor present in phenotype F4abR+/F4acR+ and a weak receptor in phenotype F4abR+/F4acR–) led us to conclude that the receptor for F4ac binds F4ab bacteria as well, and that it is controlled by one gene localized between S0068 and Sw1030 on chromosome 13.
Introduction
Enterotoxigenic Escherichia coli (ETEC) with fimbriae of the F4 (K88) family are frequently associated with diarrhoea in neonatal and in weaned pigs (Moon et al. 1999). In a large study, the mortality because of pre-weaning diarrhoea has been reported as 2.7% of the piglets born, representing 11.9% of the total mortality during that period (Nielsen et al. 1974). F4 ETEC possess proteinaceous appendages called fimbriae, which allow the bacteria to adhere to F4 variant specific receptors (Bijlsma et al. 1982) on brush borders of enterocytes and consequently, to colonize the small intestine of young pigs. Colonizing bacteria produce enterotoxins that stimulate the secretion of water and electrolytes into the lumen of the small intestine and lead to diarrhoea (Moon et al. 1999).
Three antigenic variants of F4 have been described: F4ab, F4ac and F4ad (Guinée & Jansen 1979). Nevertheless, F4ac is the only variant that has been identified in the US (Westerman et al. 1988), and the F4ac variant is dominating in Spain (González et al. 1995). In a study from Switzerland, 54 and 47%, respectively, of the Swiss Large White and Swiss Landrace pigs had the F4ac receptor phenotype (Gautschi & Schwörer 1989). Five patterns of adherence were identified by Bijlsma et al. (1982): these phenotypes were designated A (binds F4ab, F4ac and F4ad), B (F4ab and F4ac), C (F4ab and F4ad), D (F4ad) and E (binds no F4 variant). Baker et al. (1997) confirmed these five phenotypes and identified a sixth: the phenotype F (binds F4ab only). Phenotypic differences between pigs have been shown to be of genetic origin (Gibbons et al. 1977). Sellwood (1979) found evidence that the adherence is a dominant trait, inherited in Mendelian way with the two alleles: S (adhesion, susceptible) and s (non-adhesion, resistant), S being dominant over s. The loci encoding for the porcine intestinal receptors for E. coli F4ab and F4ac (ECF4abR and ECF4acR) were assigned to chromosome 13 by linkage analysis (Guérin et al. 1993; Edfors-Lilja et al. 1995). Moreover, the transferrin (TF) locus, mapped to chromosome 13 by in situ hybridization (ISH) (Chowdhary et al. 1993), is tightly linked to F4abR and F4acR (Guérin et al. 1993; Edfors-Lilja et al. 1995). The terms F4abR and F4acR refer to ECF4abR and ECF4acR throughout this paper.
In this study we refine the localization of the receptor locus for fimbriae F4ac. Pigs from a selected family were phenotyped in a microscopic adhesion test for F4ab and F4ac, and examined for markers on chromosome 13. Multipoint analysis was performed to estimate the relative position of the F4acR locus.
Materials and methods
Animals
The experimental herd was bred for this study at the Department of Farm Animals, University of Zurich. It was not representative of the general population. The pigs belonged to Large White and Large White/Landrace cross breeds. The three-generation pedigree comprised eight founders (three sires and five dams), 18 F1 and 174 F2 animals. For the linkage analysis 200 pigs in total were available. F2 pigs from matings of heterozygous susceptible (F4acR+/–) and homozygous resistant (F4acR–/–) parents were selected for segregation studies. Seventeen litters (F2 pigs) were produced from seven sires and 11 dams. A total of 187 pigs were examined in the microscopic adhesion test: they comprised 171 F2 pigs slaughtered at 8 weeks of age, three F2 fattening pigs, eight F1 parents and five founders. The remaining 13 pigs (10 parents and three founders) were kept for further production of piglets. The results of the microscopic adhesion test of the offspring were used to derive the genotypes of the parents based on the dominance hypothesis of Sellwood (1979).
Microscopic enterocyte adhesion test
The phenotype (susceptible or resistant) was established by means of a microscopic enterocyte adhesion test. The two strains of E. coli used were obtained from the Central Veterinary Laboratory, Weybridge, Surrey GB. They carried the designations E68 I (O141: K85ab: F4ab) and G4 (O45: K ‘E65’: F4ac) (Thorns et al. 1987). On the day of the test, the bacteria were grown in tryptic soy broth to early exponential phase. At an age of about 8 weeks, pigs were separated from the sow and not fed 1 day before slaughter. Preparation of enterocytes and adhesion tests were carried out as described by Vögeli et al. (1996) with the following slight modifications. Care was taken to select a jejunal segment free of contents. A single person examined the microscopic slides. Twenty well-separated and intact enterocytes were scored routinely. However, in cases with less than 20% of cells with more than five adhering bacteria, another 20 cells were examined. A pig was classified as susceptible to F4ab E. coli if at least one (2.5%) out of the 40 cells bound more than five bacteria on the brush border. For F4ac this value was set higher: a pig was classified as susceptible if more than 15% of the cells bound more than five bacteria. The different threshold values were based on F4abR and F4acR negative control matings, not included in the linkage study.
Transferrin and microsatellite polymorphisms
The TF variants were analysed using two-dimensional (Agarose-PAGE) electrophoresis techniques (Gahne & Juneja 1986). In the first dimension a 1% agarose gel (pH 5.4) and in the second dimension a 12% polyacrylamide gel (pH 8.6) were used. Microsatellites were amplified according to conditions described in the original reference (Rohrer et al. 1996). Polymerase chain reaction (PCR) was carried out in a 25-μl reaction volume containing 100 ng of porcine genomic DNA extracted from blood, 10 mm Tris-HCl (pH 9.0), 50 mm KCl, 1.5 mm MgCl2, 200 μm of each dNTP, 0.4 μm forward and reverse primers, and 1.25 U of Taq DNA polymerase (Pharmacia Biotech, Uppsala, Sweden). After an initial denaturation at 95 °C for 5 min, PCR reactions were incubated for 30 cycles of 95 °C for 30 s, 55–62 °C for 30 s, and 72 °C for 30 s. A final elongation was carried out at 72 °C for 7 min with Genescan-350 TAMRA size standard (Applied Biosystems, Perkin-Elmer Corp., Foster City, CA, USA), formamide diluted samples were analysed on a 377 ABI sequencer (Applied Biosystems). Results were evaluated with an ABI 672 Genescan program and Genotyper software (version 2.1, Applied Biosystems).
Statistics
Pairwise linkage analysis and calculation of recombination fractions was performed with CRI-MAP version 2.4 (CRIMAP, Washington Univesity, St. Louis, MO, USA) (Green et al. 1990). Multipoint linkage analysis was performed by sequential insertion of loci with option ‘ALL’. Option ‘FLIPS’ was used to determine the statistical support for the obtained order. To establish the genetic map the option ‘FIXED’ was used and the recombination fractions were converted into genetic distances using the Kosambi mapping function.
Results
Adhesion test
The results of adhesion tests with the two fimbrial variants F4ab and F4ac are summarized in Table 1. In the pedigree under study, only 38 of 171 pigs (22%), were resistant (exhibited no adhesion), whereas 93 pigs (54.5%) had both F4abR and F4acR and 40 pigs (23.5%) expressed F4abR+/F4acR–. No animals were categorized F4abR–/F4acR+. For all 509 pigs of the experimental herd tested, the percentages of the different phenotypes were similar to the pedigree used for the linkage analysis. Table 1 shows that there were two distinct tendencies in the adhesion properties of enterocytes. Pigs susceptible to F4ab and F4ac showed a strong adhesion: 81 ± 18.9% and 79 ± 17.5%, respectively, of the enterocytes bound more than five bacteria. In contrast, pigs of the phenotype F4abR+/F4acR– (susceptible to F4ab adhesion and resistant to F4ac) exhibited a weak adhesion. Only 20 ± 14.1% of the enterocytes bound more than five F4ab and less than 1% of the enterocytes bound more than five F4ac E. coli bacteria. In resistant pigs, few enterocytes with more than five bacteria were observed (0.04% for fimbriae F4ab and 0.03% for fimbriae F4ac).
| Phenotype | All tested pigs | Selected pedigree | Percent of enterocytes binding >5 bacteria of variant | |||
|---|---|---|---|---|---|---|
| Number | % | Number | % | F4ab1 | F4ac2 | |
| F4abR+/F4acR+ (F4bcR+) | 262 | 51 | 93 | 54.5 | 81 (±18.9) | 79 (±17.5) |
| F4abR+/F4acR– (F4abRw) | 117 | 23 | 40 | 23.5 | 20 (±14.1) | 0.93 (±3) |
| F4abR–/F4acR+ | 0 | 0 | 0 | 0 | 0 | 0 |
| F4abR–/F4acR– | 130 | 26 | 38 | 22 | 0.04 (±0.3) | 0.03 (±0.2) |
| Total | 509 | 100 | 171 | 100 | ||
- 1 Threshold value ≥ 2.5% enterocytes (1 of 40) with >5 adhering bacteria.
- 2 2 Threshold value >15% enterocytes (3 of 20) with >5 adhering bacteria.
Figure 1 shows the distribution of pigs in the pedigree with regard to receptor phenotype and binding strength of enterocytes. Enterocytes with more than five bacteria of either the F4ab or the F4ac variant (phenotype F4abR+/F4acR+) were predominant (70–100% of the enterocytes) in the majority of the pigs. Strong adhesion of F4ab was highly correlated with F4ac adhesion (r = 0.80). In contrast, enterocytes of the majority of the pigs being F4abR+/F4acR– exhibited a weak adhesion, as only 10–30% of the enterocytes bound more than five bacteria of the F4ab variant. We therefore designate the receptor (F4abR+/F4acR–) as a weak receptor (F4abRw). No correlation was detected between the frequencies of F4abRw and F4acR+.
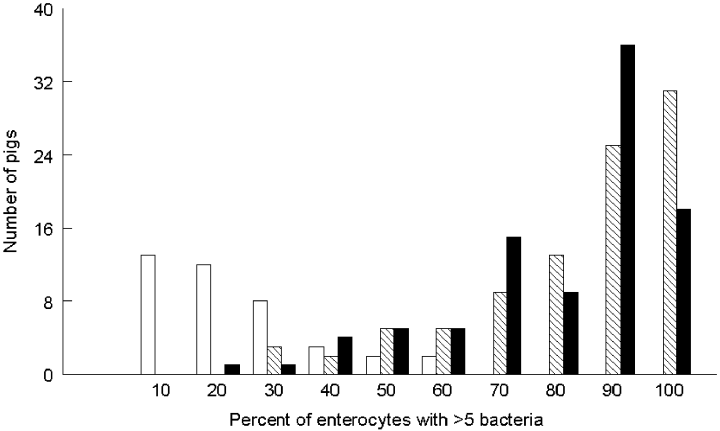
Distribution of pigs (n = 133) with enterocytes binding more than five bacteria for three receptors: F4abRw (weak receptor for F4ab detected in the phenotype F4abR+/F4acR–) (white boxes), F4abR+ (hatched boxes) and F4acR+ (black boxes) (both receptors detected in the phenotype F4abR+/F4acR+).
Linkage analysis
Pairwise lod scores and recombination fractions including F4acR and 11 marker loci are presented in Table 2. All markers were significantly linked (lod scores (Z) between 11.4 and 40.4) with F4acR. The lack of recombination detection between F4acR and Sw225 and S0075 is not because of lack of informative markers as the Polymorphism Information Contents (PIC) were, 0.60 for Sw225 and 0.61 for S0075. A PIC value higher than 0.5 indicates a high degree of polymorphism for the marker (Botstein et al. 1980).
| Marker | θ | Z |
|---|---|---|
| S0075 | 0.00 | 33.4 |
| Sw225 | 0.00 | 27.1 |
| Sw1030 | 0.01 | 34.3 |
| Sw2459 | 0.01 | 28.9 |
| Sw1386 | 0.02 | 25.5 |
| S0068 | 0.03 | 28.0 |
| Sw398 | 0.03 | 40.4 |
| Sw520 | 0.04 | 33.6 |
| TF | 0.04 | 11.4 |
| Swr1306 | 0.05 | 13.7 |
| S0222 | 0.11 | 17.9 |
Linkage analysis for F4abRw could not be performed for the following reasons. If two genes control the receptors for F4ab and F4ac, four different phenotypes would be expected. In our pedigree only three phenotypes occurred in accordance with Bijlsma et al. (1982), Edfors-Lilja et al. (1986, 1995), Rapacz & Hasler-Rapacz (1986), Guérin et al. (1993) and Peelman (1999): F4abR+/F4acR+, F4abR–/F4acR– and F4abRw. Interestingly, 40 (23.5%) from a total of 171 pigs slaughtered at 8 weeks of age for adhesion test expressed the F4abRw; and out of all pigs tested 117 (23%) displayed this phenotype. No pigs were found to display the F4abR–/F4acR+ phenotype. Consequently, two phenotypes have to be considered for the linkage analysis of F4abR: a strong (present in phenotype F4abR+/F4acR+) and a weak receptor (F4abRw) present in F4abR+/F4acR– (Table 1). To examine the transmission of F4abRw, only matings of parents lacking F4acR are informative. Owing to insufficient number of litters issued from F4ac-resistant parents, it was not possible to show the inheritance of F4abRw.
Figure 2 shows the sex-averaged Nordic. 2 map of Marklund et al. (1996) compared with the map deduced from our pedigree. The order of and the genetic distances (Kosambi cM) between F4acR and 11 markers are given. The differences in log likelihood (ΔlogL) against the inversion of adjacent loci are given to the right of the map. F4acR lay inside of the interval S0068-Sw1030, but the multipoint analysis did not allow the exact localization in the marker locus order S0068-S0075 (Sw225)-Sw1030 to be determined.
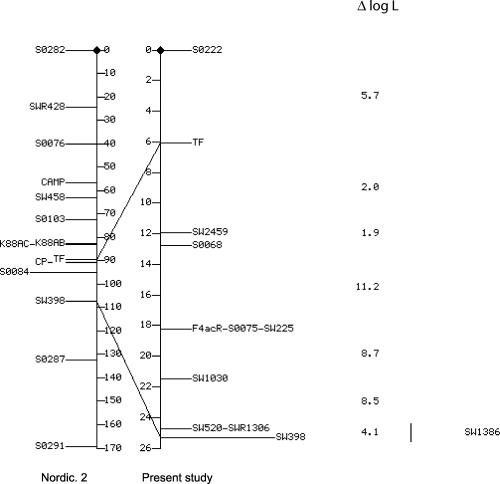
Assignment of the F4acR (K88AC) gene on porcine chromosome 13. Comparison between Nordic. 2 map (Marklund et al. 1996) and the map constructed from our data. Sex-averaged map distances are given in Kosambi cM and common loci in both maps are connected by a line. Differences in log likelihood (Δ log L) against the inversion of adjacent loci are shown to the right of the map. The order of adjacent loci supported by odds of at least 1000:1 is significant. Marker Sw1386 could not be mapped precisely. The interval of its localization is indicated by a vertical bar.
Discussion
The pigs of the phenotype F4abR+/F4acR+ showed a strong adherence: 81% of enterocytes with F4ab variant and 79% with F4ac bound more than five bacteria per cell (Table 1). For pigs expressing F4abR+/F4acR– the percentage of enterocytes with more than five bacteria was low (20%). We therefore suggest two distinct F4abR phenotypes: weak (F4abRw) and strong (F4abR). These observations are in concordance with the results of Baker et al. (1997), who noticed a poor binding of F4ab bacteria to brush borders from pigs of the phenotype F4abR+/F4acR–, in contrast to pigs of phenotype F4abR+/F4acR+. The significance of the F4abRw for the susceptibility of pigs to collibacillosis has not been determined (Baker et al. 1997). Billey et al. (1998) considered the weak binding of F4ab bacteria as an artefact, and therefore pigs susceptible to F4ab would belong to the resistant phenotype. Weak adhesion may not be detected with the same sensitivity in different adhesion tests. Because of the limited number of informative animals, we were unable to show the inheritance of F4abRw.
Sellwood (1980) described an intermediate phenotype for F4acR, which was weak-adhesive with few bacteria attaching to the brush borders. The inoculation of weak-adhesive piglets with E. coli F4ac showed that such piglets did not develop diarrhoea (Sellwood 1984; Bijlsma & Bouw 1987). Therefore, it seems that this weak receptor is less efficient at binding or there are fewer receptors per cell. We have evidence that a weak receptor for F4ac occurs in some pigs of the experimental herd not included in the selected pedigree (data not shown).
Chowdhary et al. (1993) assigned the TF gene by ISH to the q3.1 band of chromosome 13. Several studies emphasized the existence of a linkage between the F4acR locus and the serum transferrin (TF) locus and, consequently, the attribution of the F4acR to the chromosome 13 (Gibbons et al. 1977; Guérin et al. 1993; Edfors-Lilja et al. 1995). Our two-point analysis demonstrates similar linkage between F4acR and TF with a lod score of 11.4 for a recombination fraction of 0.04.
Our data suggest a different location of F4acR compared with other linkage analyses: F4acR is localized in the interval S0068-Sw1030, close to the two markers S0075 and Sw225. In the multipoint analysis, it was not possible to determine their relative position because of the lack of recombinant pigs. In contrast, Peelman (1999) assigned F4acR outside of S0068-S0075 with a recombination fraction of 0.06 to S0068, and Edfors-Lilja et al. (1995) obtained F4acR-TF-CP-S0084-Sw398 as the most likely gene order (Fig. 2: Nordic. 2 map). Our 11-point analysis indicates that the F4acR locus is located between S0068 and Sw1030, with recombination fractions (θ) of 0.05 between S0068 and F4acR, and 0.04 with Sw1030. Cytogenetic comparisons (Van Poucke et al. 1997, 1999, 2001) between human chromosome 3 and porcine chromosome 13, and linkage studies should permit the identification of candidate genes for F4acR.
A phenotype F4abR–/F4acR+ (non-adhesive for F4ab fimbriae and adhesive for F4ac) was not detected in the litters selected for the genetic analysis, nor in the remaining pigs tested. Bonneau et al. (1990) reported five pigs of 149 expressing only F4acR. In our study 23% of the pigs tested expressed F4abRw, while in other breeds studied very few pigs of that phenotype have been found (Bijlsma et al. 1982; Edfors-Lilja et al. 1986, 1995; Rapacz & Hasler-Rapacz 1986; Guérin et al. 1993; Peelman 1999). Thus it was proposed that the inheritance of F4abR and F4acR genes is under the control of two closely linked loci (θ = 0.02) (Bonneau et al. 1990; Guérin et al. 1993). Recombinant pigs (F4abR+/F4acR–, F4abR–/F4acR+) should be in the range of 2%. Our results are inconsistent with the hypothesis of two closely linked loci. In the case of F4abRw pigs we would expect a recombination frequency clearly above 2%, and in the case of F4abR–/F4acR+ pigs which were not observed we would assume a recombination frequency of zero. Based on our results as well as the literature (Billey et al. 1998), we have additional evidence that the receptor for F4ac binds F4ab bacteria as well. If F4ab and F4ac did not bind to the same receptor, a phenotype binding only F4ac should be detected. The distribution of the pigs and the binding properties of enterocytes expressing F4abR and F4acR, phenotype F4abR+/F4acR+ (Fig. 1), gives further evidence that the strong receptor for F4ab coincides with the presence of F4acR. Thus, the most probable explanation is that F4ab (strong adhesion) and F4ac bind to the same receptor, controlled by one gene that we name F4bcR.
Acknowledgements
These studies were supported by grants from the Swiss National Research Foundation (grant: 31–56613.99) and the Breeding Organization Suisseporcs (SUISAG), Sempach. We thank Dr M. Rothschild, coordinator of the US pig genome project, for providing the primers of the USDA microsatellite markers, and thank Dr M. M. Wittenbrink, Faculty of Veterinary Medicine, Zurich, for his generous support.




