Impact of Peroxisome Proliferator–activated Receptors γ and δ on Adiposity in Toddlers and Preschoolers in the GENESIS Study
Abstract
Peroxisome proliferator–activated receptor γ (PPAR γ) and peroxisome proliferator–activated receptor δ (PPAR δ) are promising candidate genes for obesity. Associations between adiposity-related phenotypes and genetic variation in PPAR γ (Pro12Ala and C1431T), as well as PPAR δ (T+294C) were assessed in 2,102 Greek children aged 1–6 years, as part of a large-scale epidemiological study (Growth, Exercise and Nutrition Epidemiological Study In preSchoolers). In girls aged 3–4 years, the Ala12 allele was associated with higher mid-upper arm (P = 0.010) and hip (P = 0.005) circumferences, as well as subscapular (P = 0.008) and total skinfolds (P = 0.011) that explained 2.0, 3.7, 2.1, and 1.9% of the phenotypic variance, respectively, while the T1431 allele was associated with higher mean values for waist circumference (P = 0.018) and suprailiac skinfold (P = 0.017), genotype accounting for 1.6% of the variance in both phenotypes. No significant effects of PPAR δ T+294C polymorphism or the interaction of the PPAR δ and PPAR γ variants on adiposity-related phenotypes were observed in any age group or gender. Haplotype-based analysis including both PPAR γ polymorphisms revealed that in girls aged 3–4 years, the Ala-T haplotype was associated with higher waist (P = 0.014) and hip (P = 0.007) circumferences compared to the common Pro-C haplotype. The PPAR γ Pro12Ala and C1431T polymorphisms are associated with increased adiposity during early childhood in a gender- and age-specific manner and independently of the PPAR δ T+294C polymorphism.
The prevalence of overweight and obesity in children in the developed world has reached epidemic proportions (1). Twin and family studies have shown variable estimates of heritability for obesity and fat deposition ranging from 40 to 70% in different studies supporting the idea of a strong genetic component in the development of obesity (2). However, other studies have indicated that changes in key environmental factors, such as diet and levels of physical activity, are responsible for this increase (3), and gene-environment interactions can exert a variable effect on adiposity with age (2). Among the 127 genes currently identified in obesity association studies are the peroxisome proliferator–activated receptor γ (PPARγ) and peroxisome proliferator–activated receptor δ (PPARδ) genes (4). PPARγ and PPARδ are transcriptional factors that control transcription of target genes linked to energy metabolism and homeostasis and are activated by both dietary fatty acids and their metabolic derivatives (5). PPARγ and PPARδ are both expressed in adipose tissue, where the former has an important role in adipocyte differentiation, as well as lipid and glucose homeostasis, and the latter is involved in fatty-acid oxidation and energy uncoupling (5).
Two common polymorphisms, a proline (Pro) to alanine (Ala) substitution located at codon 12 (Pro12Ala) (6) and a synonymous C to T substitution in exon 6 at nucleotide 1431 (C1431T) (7) have been detected in the PPARγ gene. Both the Pro12Ala and C1431T polymorphisms have been investigated for their association with obesity and type 2 diabetes, with Ala12 and T1431 carriers showing higher and lower BMI, respectively (for review, see ref. 8). In the PPARδ gene, one common polymorphism, T+294C, was detected in exon 4 of the 5′ untranslated region (9). In middle-aged men, the C+294 allele was associated with higher plasma low density lipoprotein cholesterol and showed higher transcriptional activity compared to the T+294 allele (9), while in moderately obese and dyslipidemic middle-aged men and women a trend toward an interactive effect between the PPARδ and PPARγ genotypes on BMI was observed (10). To investigate the potentially higher genetic contribution in a younger population, this study aimed to assess associations between PPARγ and PPARδ polymorphisms and adiposity-related phenotypes in a sample of Greek toddlers and preschoolers (Growth, Exercise and Nutrition Epidemiological Study In preSchoolers) (11).
In the final analysis, 2,102 children (1,095 boys and 1,007 girls) were included, after excluding subjects with missing phenotypic data or no DNA sample. Pro12Ala and C1431T genotypes were determined for 1,888 (981 boys and 907 girls) and 1,956 (1,019 boys and 937 girls) individuals, respectively. Genotyping for T+294C polymorphism was successful in 1,962 individuals (1,015 boys and 947 girls). The population was in Hardy–Weinberg equilibrium and the frequencies of the minor alleles were f(Ala12) = 0.07, f(T1431) = 0.09, and f(C+294) = 0.20. The frequency of the Ala12 was lower than that reported in French adults (0.11) (12) but similar to that reported in Greek school-age children (0.07) (13) and in adults from Southern Europe (0.07) (14). Similarly, the frequency of the T1431 variant was also lower than that reported for the French cohort (0.13) (12), whereas the frequency of the C+294 allele was similar to that reported in French-Canadian adults (0.20) (15). The diplotype distribution for the PPARγ polymorphisms, as well as the frequencies of the inferred haplotypes, are shown in Table 1.
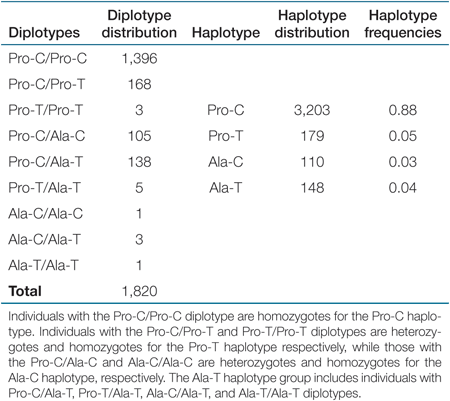 |
Genotypic associations between the PPARγ and PPARδ genotypes and adiposity-related phenotypes were assessed by ANOVA, as described in “Research Methods and Procedures.” Full analyses are given in Supplementary Tables S1–S6 online. After performing the Dunn–Sidak correction for multiple testing for each age group, statistically significant (α value = 0.020) differences were observed between the means of the Pro12Ala genotypes for mid-upper arm (P = 0.010) and hip (P = 0.005) circumferences, as well as subscapular skinfold (P = 0.008) in girls aged 3–4 years, and these associations accounted for 2.0, 3.7, and 2.1% of the phenotypic variance, respectively (Table 2). In the same age group, a Sidak correction for multiple testing revealed significant (α value = 0.020) differences between the means of C1431T genotypes for waist circumference (P = 0.018) and suprailiac skinfold (P = 0.017) explaining 1.6% of the variance in both phenotypes (Table 2). ANOVA tests for associations between the sum of skinfolds and the two PPARγ polymorphisms revealed that girls aged 3–4 years carrying the Ala12 (P = 0.011) and T1431 (P = 0.045) alleles had higher sum of skinfolds, but only the association with the Ala12 variant was statistically significant after Sidak correction, accounting for 1.9% of the phenotypic variance (Table 2). A significant association between the T+294C polymorphism and hip circumference (P = 0.009) was observed in girls aged 1–2 years with carriers of the +294T allele, showing higher adiposity. However, given the fact that no other significant associations were observed in the same age group, and as such there was no consistency across the phenotypic groups, it is possible that this association is most probably a type 1 error given also the small number of subjects in this age group.
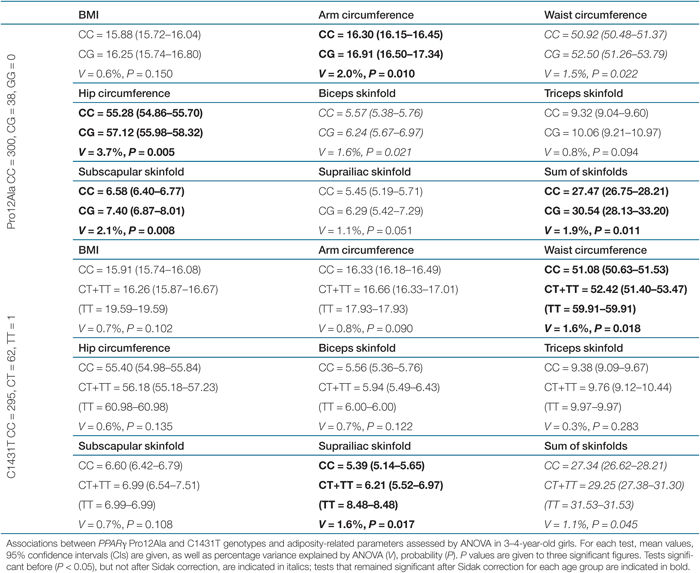 |
Associations significant after correction for multiple testing were further assessed by haplotype-based analysis. In girls aged 3–4 years, ANOVA tests revealed differences between the PPARγ haplotypes and the mean values of waist (P = 0.035) and hip (P = 0.038) circumferences, as well as suprailiac skinfold (P = 0.039) (Table 3). The extra portion of the variance accounted for using diplotypes instead of individual genotypes was not significant for any phenotype, as indicated by the diplotype improvement (Table 4). Postcomparisons between diplotypes for the aforementioned phenotypes revealed that the Ala-T haplotype was associated with significantly higher waist (P = 0.014) and hip (P = 0.007) circumferences compared to the common Pro-C haplotype, whereas the Ala-C haplotype showed significantly higher scores for suprailiac skinfold (P = 0.016) compared to the Pro-C haplotype (Table 3, 1). A comprehensive analysis of PPARγ haplotypes is shown in Supplementary Tables S7 and S8 online. No interactive effect between the PPARγ and PPARδ gene polymorphisms on adiposity-related phenotypes was observed (data not shown).
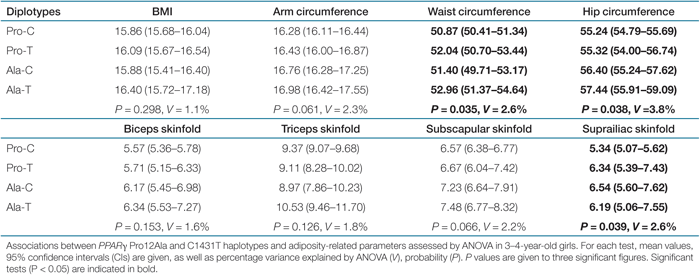 |
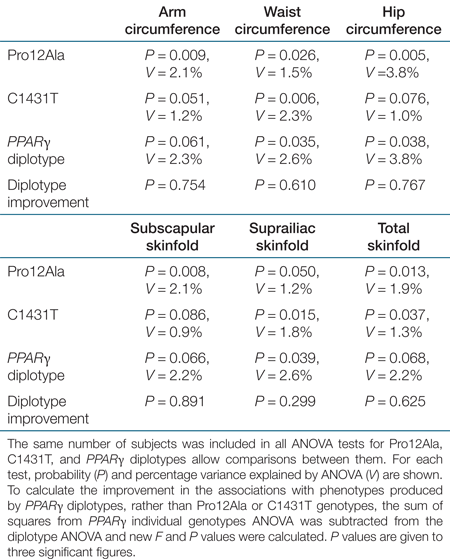 |
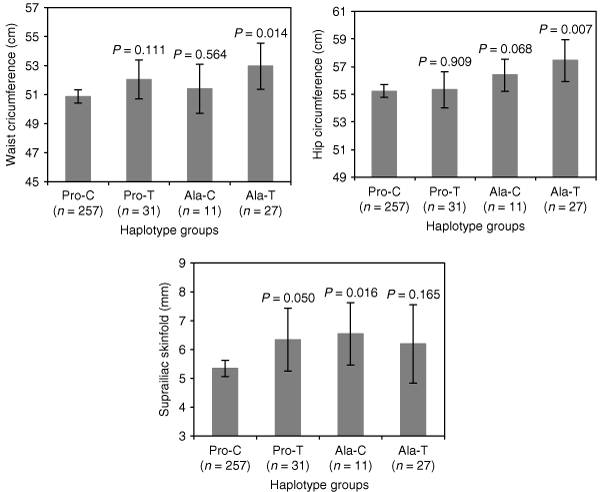
: Mean waist and hip circumferences, as well as suprailiac skinfold within each haplotype group in girls aged 3–4 years. The s.e.m. for these phenotypes are also indicated. Mean values within the Pro-C haplotype group for these phenotypes were compared to those in Pro-T, Ala-C, and Ala-T groups and P values are given to three significant places.
The significant associations of the Ala12 and T1431 variants with higher adiposity reported in this study are in accordance with previous studies, albeit in overweight adults but contrary to other studies where the Ala12 allele was associated with lower BMI (for review, see ref. 8). Although the PPARδ C+294 allele has been previously associated with lower BMI (10), no significant associations between the T+294C polymorphism and adiposity-related phenotypes were observed in this and previous studies (16). Haplotype-based analysis for the PPARγ polymorphisms did not reveal stronger associations than genotype-based analysis, and the portion of variance explained by the diplotypes, although generally higher than that explained by individual genotypes (based on the increase in the sum of squares and as a result of the increased degrees of freedom) (Table 4), was not significantly larger, suggesting that most of the genetic effect observed is accounted for either by the individual single-nucleotide polymorphisms or by variants which are in strong linkage disequilibrium with these single-nucleotide polymorphisms. However, haplotype-based analysis enabled the significant associations of the Ala12 and T1431 alleles with increased adiposity to be confirmed, since the Ala-T haplotype was observed to have an augmented effect on adiposity compared to the Pro-T and Ala-C haplotypes, which assessed the effect of T1431 and Ala12 independently of Ala12 and T1431 alleles, respectively (1). The lack of significant differences between the Pro-C and Pro-T or Ala-C haplotypes (as shown in the postcomparisons, 1) could reflect the absence of any interaction between the Pro12Ala and C1431T polymorphisms.
In this (Table 2) and in previous studies in children (17), the Pro12Ala accounted for a smaller proportion of the variance in BMI compared to that in adult populations (1% of the total variance of BMI) (18). This is possibly due to BMI being a proxy measurement of adiposity in childhood. Furthermore, the interaction of the PPARδ with genetic (10) and environmental (15) factors may exert variable effects on adiposity with age, since significant associations were detected in previous studies in adults (10,15) but not in the present population. A potential gene–gene interaction between the PPARδ T+294C and PPARγ Pro12Ala or C1431T polymorphisms was also studied; however, no interactive effect was observed between these polymorphisms (data not shown), as also shown in a previous study (10), indicating that the PPARγ polymorphisms influence adiposity independently of the T+294C variant.
In this study, significant associations between PPARγ variants and adiposity-related phenotypes were identified only in girls aged 3–4 years. This finding could suggest gender- and age-specific effects of these variants in early childhood. Gender-specific effects of the selected variants on adiposity has also been demonstrated in a previous report (19) and may be the result of the dependency of certain genetic determinants on other cofactors such as hormones or other genes for their function. Nevertheless, it has been shown that the expression of both PPAR γ1 and PPAR γ2Isoforms is sexually dimorphic, with both of them being more expressed in women than in men and the ratio of PPAR γ2 to PPAR γ1Is correlated with BMI (20). This could suggest that the effect of variation in this gene would be more pronounced in women than in men, and this is in agreement with the association of the Pro12Ala and C1431T with higher adiposity only in girls but not in boys in the present population. Furthermore, this study offers support to previous suggestions that the effect of genes may be age-specific and dependent on their age-related variability in exposure to environmental factors that predispose to their expression (2). A previous study that assessed the longitudinal effects of an ADRB2 genetic variant on adiposity from childhood to adulthood has detected no genetic effects on obesity in childhood but significant associations in adulthood (21). Given the fact that PPAR γ activators are mainly fatty acids derived from diet, differences in the dietary composition and especially fat intake between age groups could influence the potential significant effects of PPAR γ on adiposity.
Functional studies have shown that the Ala12 PPARγ receptor displays reduced DNA-binding affinity and modestly impaired transcriptional activity in target genes (22). As such, the Ala variant would be expected to protect against increased adiposity due to less efficient stimulation of target genes. However, in the present and previous cross-sectional and meta-analysis studies (18), the Ala12 allele was associated with increased adiposity (Table 2). This has been postulated to be due to the Ala12 variant enhancing somehow the antilipolytic action of insulin, which leads to reduced release of free fatty acids (23). Furthermore, although C1431T is a silent polymorphism, it was found to be a better predictor of fasting insulin levels and insulin sensitivity than the Pro12Ala (19), suggesting that the C1431T polymorphism may be in tight linkage disequilibrium with a functional variant in the PPARγ or nearby gene.
In conclusion, this study revealed that Pro12Ala and C1431T polymorphisms in PPARγ are associated individually with higher adiposity measurements in an age- and gender-specific manner and independently of the PPARδ polymorphism in early childhood. Although no interaction was observed between these two PPARγ polymorphisms, the occurrence of the two rare alleles in the same haplotype seemed to have an augmented effect on adiposity.
Research Methods and Procedures
The study was approved by the Ethics Committee of Harokopio University of Athens and by all municipalities taking part in the study. Written informed consent for the participation of their children in this study was obtained from all parents.
Subjects
The sample population has previously been described in detail (24). Subjects were selected from a large-scale epidemiological study (Growth, Exercise and Nutrition Epidemiological Study In preSchoolers) (11). Briefly, the selected cohort comprised 2,374 healthy children aged 1–6 years attending public and private nurseries in five geographical districts of Greece: Athens, Aitoloakarnania, Thessaloniki, Halkidiki, and Helia. These counties are widely scattered across Greece and account for ∼70% of the total Greek population. The sampling of the nurseries was random, multistage, and stratified by the total population of children. The study was approved by the Ethics Committee of Harokopio University of Athens and by all municipalities taking part in the study. There was no selection of subjects for inclusion based on any phenotypic characteristic, including health grounds, and therefore the study sample represents an unselected cross section of the general population in the age range 1–6 years.
Protocols for phenotypic measurements
Details of the adiposity-related phenotype measurements obtained have previously been described (11,24). Body composition measurements included weight, height, waist, hip, and mid-upper arm circumferences, as well as measurement of body fat at four sites (biceps, triceps, subscapular, and suprailiac). Weight and height were converted to BMI using Quetelet's Index (weight (kg)/height2 (m2)). Total skinfolds were estimated as the sum of the four individual skinfolds.
DNA extraction and genotype determination
DNA extraction was carried out as previously described (23). DNA amplification was carried out for two PPARγ polymorphisms, Pro12Ala (rs1801282) and C1431T (rs3856806) and one PPARδ polymorphism, T+294C (rs2016520). Primer design was based on the National Center for Biotechnology Information reference sequences NM_015869.3 and NT_007592.14 for PPARγ and PPARδ polymorphisms, respectively. Genotypes for all polymorphisms were determined by polymerase chain reaction amplification and restriction fragment length polymorphism analysis. For the Pro12Ala polymorphism, the forward primer 5′-TGTCTTGACTCATGGGTGTATTC-3′ and the reverse primer 5′-ATCAGTGAAGGAACCGCTTT-3′ were used. The reverse primer used for Pro12Ala amplification contained a mismatched base (underlined) that created a BslI restriction site in conjunction with the Pro12 allele. Thermocycling conditions consisted of 5 min at 94 °C, followed by 35 cycles at 94 °C for 45 s, 58 and 72 °C each for 30 s, and completed with 10 min at 72 °C, before being held at 4 °C until further processing. For restriction fragment length polymorphism analysis of this polymorphism, polymerase chain reaction products (5 μl) were digested overnight at 55 °C with 4 U of BslI restriction enzyme (New England Biolabs UK, Hitchin, Hertfordshire, UK). The digested fragments were resolved on a 3.5% agarose gel. The Pro12 allele (C-allele) contained a unique restriction site for BslI yielding two products of 168 base pair (bp) and 17 bp, while the Ala12 allele (G-allele) yielded an uncut 185-bp fragment.
For the C1431T polymorphism amplification, the forward primer 5′-TGAATGTGAAGCCCATTGAA-3′ and the reverse primer 5′-GAGCGGGTGAAGACTCATGT-3′ were used. Thermocycling conditions consisted of 5 min at 94 °C, followed by 35 cycles at 94 °C for 30 s, 57 and 72 °C each for 30 s, and completed with 10 min at 72 °C, before being held at 4 °C until further processing. For restriction fragment length polymorphism analysis, each polymerase chain reaction product (5 μl) was digested overnight with 12 U of PmlI restriction enzyme (New England Biolabs UK, Hitchin, Hertfordshire, UK), 10× Nebuffer and 1% bovine serum albumin in a total volume of 10 μl at 37 °C to obtain optimal digestion. The digested fragments were separated on 3% agarose gel. The C1431 allele (C-allele) contained a unique restriction site for PmlI, producing two fragments of 146 and 57 bp, while the T1431 (T-allele) yielded an uncut 203-bp fragment.
For the T+294C polymorphism amplification, the forward primer 5′-GCACCATCCTCTTCCTTGTC-3′ and the reverse primer 5′-CCTCTGCCACTTCCTCTTTCT-3′ were used. Thermocycling conditions consisted of 5 min at 94 °C followed by 35 cycles at 94 °C for 45 s, 58 °C for 30 s, and 72 °C for 30 s, completed with 10 min at 72 °C, before being held at 4 °C until further processing. For restriction fragment length polymorphism analysis of this polymorphism, each polymerase chain reaction product (15 μl) was digested at 55 °C for 4 h with 3 U of BslI restriction enzyme (New England Biolabs UK, Hitchin, Hertfordshire, UK). The T+294 allele (T-allele) contained a restriction site for BslI yielding two products of 201 bp and 28 bp, while the C+294 allele (C-allele) had two restriction sites yielding three fragments of 119, 82, and 28 bp. The digested fragments were separated on 2.5% agarose gel.
Data analysis
Individuals with missing phenotypic data or no DNA sample were excluded from the statistical analysis. Allele frequencies were calculated for both PPARγ and PPARδ polymorphisms, and χ2 tests were performed to assess whether the observed genotype frequencies were in Hardy–Weinberg equilibrium. The inferred haplotype distributions were calculated using the Haplotype resolution V3.0 website (http:research.calit2.nethapInstructions.htm). Subjects were analyzed separately by gender to determine the gender-specific effects of the PPARγ (25,26) and PPARδ (15) gene polymorphisms on the measured phenotypes.
Subjects from each gender were also assigned to single-year age groups: 13–24 months (1–2 years), 25–36 months (2–3 years), 37–48 months (3–4 years), and 49–71 months (4–6 years) to account for marked changes in body composition during the first decade of life (27) and to control for secular trends in adiposity (11). Phenotypic distributions for each gender and age group were tested separately for normality using the Ryan–Joiner test (28) and datasets with nonnormal distributions were transformed by Box–Cox transformation (29). Mean values and 95% confidence intervals for each phenotype were subsequently back-transformed for presentation purposes. The influences of both PPARγ and PPARδ polymorphisms on adiposity-related phenotypes were evaluated by one-way ANOVA using MINITAB 13.30 (Minitab, Coventry, UK). Because of the small number of homozygotes for the less common allele at each locus in PPARγ and PPARδ genes (i.e., Ala12, T1431 and C+294), these were collapsed with the heterozygotes and compared to the homozygotes for the common alleles for all further analyses, as previously suggested (10,25,30). The critical P value at which significance was accepted (the α value) was corrected for multiple testing (within each gender and age group) for each polymorphism using the Dunn–Sidak method, with correction for repeated tests and correlations between datasets, as implemented on the Simple Interactive Statistical Analysis website (31). For each age group, the mean correlation (r) was calculated as the square root of the mean of the squared partial correlations (coefficient of determination) between the assessed adiposity-related phenotypes.
For haplotype-based analysis, individuals with the Pro-C/Pro-T diplotype were combined with those homozygotes (Pro-T/Pro-T) for the Pro-T haplotype, as previously suggested (31). Furthermore, individuals heterozygous (Pro-C/Ala-C) and homozygotes (Ala-C/Ala-C) for the Ala-C haplotype were also collapsed into a single group, whereas double heterozygotes (Pro-C/Ala-T) were combined with individuals heterozygotes (Pro-T/Ala-T, Ala-C/Ala-T) and homozygotes (Ala-T/Ala-T) for the Ala-T haplotype (31). Differences in the phenotypic means of these four diplotype groups were assessed by one-way ANOVA for each age group and gender. Significant associations (P < 0.05) were further analyzed by performing two-tailed t-tests, which allowed the comparison of Pro-T, Ala-C and Ala-T haplotypes with the common Pro-C haplotype and the detection of any possible interactions between these two PPARγ polymorphisms. To analyze the effects of interactions between the PPARγ Pro12Ala or C1431T polymorphisms with the PPARδ T+294C polymorphism on adiposity, individuals were classified into four groups taking into consideration both PPARγ and PPARδ variants—individuals having both common alleles (Pro12 or C1431 and C+294 alleles), only one rare variant in PPARγ (Ala12, T1431) or PPARδ (T+294) genes and rare alleles in both genes. Differences in the phenotypic means of these four PPARγ/PPARδ genotype groups were assessed by one-way ANOVA.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http:www.nature.comoby
DISCLOSURE
Y. Manios also works as a scientific consultant for Friesland Food Hellas. None of the other authors declared any conflict of interest.
Acknowledgments
This research was supported in part by Grant 17/D17566 from the Medical Research Council/Biotechnology and Biological Sciences Research Council associate program in human nutrition research and a grant from Friesland Food Hellas. The sponsors had no role in the study design, collection, analysis or interpretation of the data, the writing of the manuscript, and the submission and revision of the paper. The cooperation of all schools and subjects is greatly appreciated.





