The GDI1 genes from Kluyveromyces lactis and Pichia pastoris: cloning and functional expression in Saccharomyces cerevisiae
Abstract
The nucleotide sequences of 2.8 kb and 2.9 kb fragments containing the Kluyveromyces lactis and Pichia pastoris GDI1 genes, respectively, were determined. K. lactis GDI1 was found during sequencing of a genomic library clone, whereas the P. pastoris GDI1 was obtained from a genomic library by complementing a Saccharomyces cerevisiae sec19-1 mutant strain. The sequenced DNA fragments contain open reading frames of 1338 bp (K.lactis) and 1344 bp (P. pastoris), coding for polypeptides of 445 and 447 residues, respectively. Both sequences fully complement the S. cerevisiae sec19-1 mutation. They have high degrees of homology with known GDP dissociation inhibitors from yeast species and other eukaryotes. The GenBank Accession Nos of the sequences are AF255332 (K.lactis GDI1) and AY007574 (P. pastoris GDI1). Copyright © 2001 John Wiley & Sons, Ltd.
Introduction
Vesicle targeting and fusion events in eukaryotic cells require strict regulatory mechanisms. Small GTPases of the Rab family, first identified in the yeast Saccharomyces cerevisiae (Salminen and Novick, 1987) are such regulatory factors. They cycle between the GDP-bound inactive and GTP-bound active form. The GDP dissociation inhibitors (GDIs) regulate Rab proteins by maintaining them in their inactive, soluble form in the cytosol (Garrett et al., 1994; Ullrich et al., 1993). GDI proteins are highly homologous within a wide variety of organisms including Homo sapiens, S.cerevisiae, Arabidopsis thaliana and others (Z̆árský et al., 1997). Functional similarity has been demonstrated, for example, by the ability of a cDNA coding for A. thaliana Gdi to complement the sec19-1 mutation (allelic to gdi1) of S. cerevisiae (Z̆árský et al., 1997) and by in vitro experiments with yeast Sec4p and Drosophila or bovine GDI (Garrett et al., 1993). The crystal structure of bovine GDI has been determined (Luan et al., 2000; Schalk et al., 1996). The protein has been shown to consist of two domains. Domain I ofS.cerevisiae Gdi1p binds to Rab GTPase (Luanet al., 1999; Schalk et al., 1996; Wu et al., 1998), whereas Domain II regulates membrane attachment and release of Rab (Gilbert and Burd, 2000).
In clinical studies, it has also been shown thatmutations in the human GDI1 gene coding forα-GDI lead to mental retardation (D'Adamo et al., 1998), and this emphasizes the importance ofthe role of GDI proteins as components ofcellular regulation from protein secretion inyeast to neuronal development and action in man.
In this paper we report on the complete nucleotide sequences of two genes coding for GDI proteins. The deduced amino acid sequences of the genes from Kluyveromyces lactis and Pichia pastoris show identities of 81% and 72%, respectively, when compared to S. cerevisiae Gdi1p.
Materials and methods
Strains and media
The P. pastoris strain GS115 (his4) was used as the DNA source for the P. pastoris genomic library (Payne et al., 2000) and PCR amplification of the P.pastoris GDI1 open reading frame (ORF). The K. lactis strain CBS 2359 (Kiers et al., 1998), a generous gift from Hiroshi Fukuhara (Institut Curie, Orsay, France), was used as the DNA source for the K. lactis genomic library and PCR amplification of the K. lactis GDI1 ORF. Complementation experiments were carried out with the S.cerevisiae gdi1 strain NY1213 (MATα sec19-1 ura3-52 leu2-3,112), kindly provided by Peter Novick (Department of Cell Biology, Yale University School of Medicine). The S. cerevisiae wild-type GDI1 ORF was obtained by PCR from the strain DNY1 (Nelson et al., 1993). Yeast cells were grown either in YPD medium (2% glucose, 2% peptone, 1% yeast extract) or SCD medium lacking uracil (Sherman et al., 1983). Selection against expressed URA3 was done on SCD plates containing 0.2% 5-fluoro-orotic acid (5-FOA). Liquid cultures were grown at 30°C. Plates were incubated at either 24°C, 35°C, 37°C or 38°C. The Escherichia coli strain DH5α was used as the cloning host andgrown at 37°C in LB medium, supplemented with 100 µg/ml ampicillin for selection of transformants.
DNA methods
Standard recombinant DNA methods were used (Sambrook et al., 1989). The S. cerevisiae GDI1 ORF was obtained by PCR using the primers 5′-GCATTGGATCCATGGATCAAGAAACAATAGACACT-3′ and 5′-GCATTAAGCTTTTACTGCTTTTCTTGTTCTTGTCT-3′, yielding a fragment that was digested with BamHI–HindIII and cloned into the vector YCpMET to obtain YCpM–GDI1. The plasmid YCpMET is derived from pGFP–C-FUS (Niedenthal et al., 1996) by removing by SalI digestion the GFP-coding region from this URA3-containing centromeric plasmid, which harbours the MET25 promoter. The K. lactis GDI1 ORF was obtained by PCR using the primers 5′-GCATTGGATCCATGGACGAAAATTATGATGTTATC-3′ and 5′-GCATTAAGCTTTTACTCCTCTGCAACATCTTCTCT-3′, yielding a fragment that was digested with BamHI–HindIII and cloned into YCpMET to obtain YCpM–KlGDI1. The P. pastoris GDI1 ORF was obtained by PCR using the primers 5′-GCATTGAATTCATGGAAGAAAATTACGACGTTATT-3′ and 5′-GCATTCTCGAGCTACTCACAGGCGAGGGATTCCTC-3′, yielding a fragment that was digested with EcoRI–XhoI andcloned into YCpMET to obtain YCpM–PpGDI1. Initial sequencing of the library clones was performed using in vitro transposon insertion technology (Template Generation System, Finnzymes, Finland). Sequences of the initial clones and all PCR products were verified and completed by sequencing both strands, using primers annealing at approximately 300 bp intervals. Sequencing was done using an ABI Prism 310 sequencer (PE/Applied Biosystems). Sequence assembly was done using the program DNAMAN 4.1 for Windows (Lynnon Biosoft, Canada). Sequence alignment was performed using ClustalX 1.8 (Thompson et al., 1997) and GeneDoc 2.6.001 (http://www.psc.edu/biomed/genedoc/, accessed December 2000). Transformation of S. cerevisiae cells was performed by a modified lithium acetate method (Toikkanen et al., 1996).
Construction of the K. lactis genomic library
Genomic DNA from the K. lactis wild-type strain CBS 2359 was isolated and partially digested by the Sau3A restriction enzyme. The digested fragments were size-selected in a sucrose gradient and purified using standard methods (Campbell and Duffus, 1988; Sambrook et al., 1989). The DNA fragments were ligated into the polylinker BamHI site of the yeast multicopy vector YEplac181 (Gietz and Sugino, 1988). The ligation mixture was transformed into E. coli and the resulting 46 000 independent clones were collected and pooled. 30% of the clones contained an insert with an average size of 1.7 kb.
Nucleotide sequence Accession Nos
The Genbank/EMBL Accession Nos for the sequences reported in this paper are AF255332 (K.lactis) and AY007574 (P. pastoris).
Results and discussion
Cloning of the Kluyveromyces lactis GDI1 gene
The K. lactis GDI1 gene was found during sequencing of a library clone obtained from a screening not related to this work. Initial testing of functionality was done by introducing the library clone into the sec19-1 mutant strain, in which it suppressed the temperature-sensitive phenotype. Full complementation at both 37°C and 38°C was also achieved with the centromeric plasmid YCpM–KlGDI1 containing the K. lactis GDI1 ORF obtained byPCR. A genomic fragment of 2794 bp was sequenced (Figure 1). It contains the ORF of 1338 bp, coding for a protein of 445 amino acidswith a calculated molecular weight of 50.3 kDaand a pI value of 5.4 (http://www.expasy.ch/cgi-bin/pi_tool). No intron was found. On the opposite strand of the promoter region, at position −511, another potential ORF starts. The partial sequence has its highest homology with the putative D-2-hydroxyacid dehydrogenase YGL185c of S. cerevisiae.
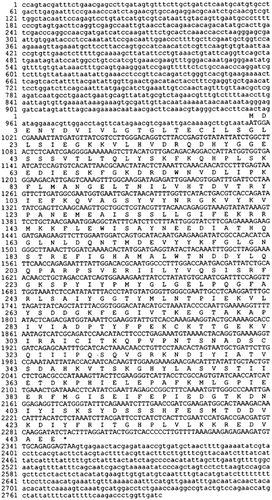
Nucleotide and deduced amino acid sequences of the K. lactis GDI1 gene. The amino acid sequence is given above the nucleotide sequence of the GDI1 coding region, shown in upper case letters
Cloning of the Pichia pastoris GDI1 gene
To obtain the P. pastoris GDI1 gene, we used the P.pastoris genomic library to transform the sec19-1 temperature-sensitive yeast strain. After the transformation, cells were plated on selective medium and incubated at 24°C for 16 h to allow gene expression to start. Plates were then shifted to the restrictive temperature of 35°C and incubated for a further 2 days. Approximately 2 million transformants were screened on 28 plates, yielding ca. 1000 temperature-resistant colonies. 144 Colonies were picked at random, patched on YPD plates and grown overnight at 35°C. The patches grown on YPD were replicated onto plates containing 5-FOA and incubated for 3 days at 35°C. Patches showing papillar growth were discarded as originating from possible revertants not needing the URA3-containing library plasmid for survival. 16 patches showing no growth were selected, and the plasmids were extracted and transformed into E. coli for preparation of DNA. Retransformation into the sec19-1 strain yielded two library plasmids that suppressed the temperature-sensitive phenotype. From both plasmids the P. pastoris GDI1 gene was identified by sequencing. Full complementation at both 37°C and 38°C was also achieved with the centromeric plasmid YCpM–PpGDI1 containing the P. pastoris GDI1 ORF amplified by PCR. In total, a genomic fragment of 2876 bp was sequenced (Figure 2). It contains the ORF of 1344 bp coding for a protein of 447 amino acids with a calculated molecular weight of 50.7 kDa and pI value of 5.4. No intron was found. The opposite strand of the terminator region contains a potential ORF starting at position 2870. The possibly partial ORF codes for215 amino acid residues with a weak similarity to S.cerevisiae Bas1p and other transcription factors.
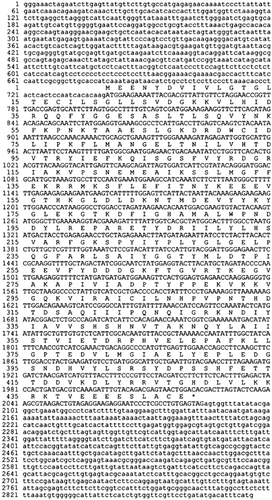
Nucleotide and deduced amino acid sequences of the P. pastoris GDI1 gene. The amino acid sequence is given above the nucleotide sequence of the GDI1 coding region, shown in upper case letters
Complementation of the Saccharomyces cerevisiae sec19-1 temperature-sensitive mutation
To test the functionality of the cloned K. lactis and P. pastoris GDI1 genes and to perform a semi-quantitative comparison of functionalities of the cloned genes with the wild-type S. cerevisiae GDI1 gene, all three were cloned into YCpMET under the control of the MET25 promoter. The medium contained 1 mM methionine, which causes the cloned genes to be expressed on the weak basal level (Mumberg et al., 1994; Sangsoda et al., 1985). These plasmids and a vector control were introduced into the sec19-1 strain, and equal amounts and dilutions of cell suspensions were spotted onto selective plates at 24°C, 37°C and 38°C (Figure 3). Both the K. lactis and the P. pastoris GDI1 genes were able, and in a very similar way to the S. cerevisiae GDI1, to fully complement the temperature-sensitive phenotype of the sec19-1 strain NY1213. This is clearly attributable to the high homology between Gdi proteins found in various eukaryotic organisms (Figure 4).
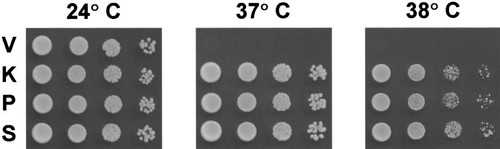
Complementation analysis of the sec19-1 strain done on SCD −ura plates. Plates were spotted with 3 µl 10-fold serial dilutions of cells (OD600: 1, 0.1, 0.01, 0.001) transformed with the vector control YCpMET (V), with YCpM–KlGDI1 containing the K. lactis GDI1 ORF (K), with YCpM–PpGDI1 containing the P. pastoris GDI1 ORF (P) or with YCpM–GDI1 containing the S. cerevisiae GDI1 ORF (S). The plates were incubated for 3 days at 24°C, 37°C and 38°C, respectively

Comparison of the deduced amino acid sequences of the identified GDI1 ORFs with other homologues. Amino acid identity is denoted by a black background. Sequences shown are human Rab GDI alpha (HsGdi, Accession No. BAA08078), Drosophila melanogaster GDI (DmGdi, AAA28567), Caenorhabditis elegans GDI (CeGdi, AAA17051), Arabidopsis thaliana GDI (AtGdi, CAA69258), Saccharomyces cerevisiae Gdi1p (ScGdi, AAB30540), Kluyveromyces lactis GDI (KlGdi, AF255332), Pichia pastoris GDI (PpGdi, AAG12984) and Schizosaccharomyces pombe GDI (SpGdi, T38215)
Acknowledgements
We thank Benjamin S. Glick (University of Chicago) for kindly providing the P. pastoris genomic library. We are grateful to Outi Könönen for excellent technical assistance. The work was financially supported by the Academy of Finland (Grant No. 49894 to S.K.), The Finnish Centre of Excellence Programme. Part of the support for, M.H.B. came from the Viikki Graduate School in Biosciences.




