Disentangling relationships in Euphorbia agraria s.l. (Euphorbiaceae) in south-east Europe: One or two species?
Associate Editor: Natascha Dorothea Wagner
Abstract
We explored the diversification of the south-eastern and eastern European Euphorbia agraria s.l. applying molecular (amplified fragment length polymorphism [AFLP] fingerprinting and sequencing of the nuclear ribosomal internal transcribed spacer [ITS]), cytogenetic (relative genome size [RGS] estimations) and morphometric methods. The AFLP data, and to a lesser extent ITS sequences, inferred two main phylogenetic lineages corresponding to eastern Balkan-Pontic E. agraria and central Balkan E. subhastata; their closest relatives are E. tommasiniana endemic to the north-westernmost Balkan Peninsula and E. salicifolia, which is more widespread in the Balkan Peninsula. Genetic divergence is reflected in morphological and ecological differentiation, rendering recognition of E. subhastata – previously segregated as a variety – at the species level, resulting in a revised taxonomic treatment. Euphorbia agraria thrives in grasslands and ruderal places that are widespread in the lowlands of the eastern Balkan Peninsula and the adjacent Pontic region; the continuity of the habitat probably confers weak genetic differentiation within this species. On the other hand, E. subhastata grows in screes and open forests of river gorges separated by mountain ridges, leading to more pronounced inter-population differentiation. The RGS data revealed di- and tetraploid populations within both species and in combination with phylogenetic results suggest recurrent autopolyploidisation. Our results support the Balkan Peninsula as a hotspot of genetic and species diversity and indicate that future biodiversity research in this part of Europe should focus on the central, eastern and southern Balkan Peninsula that were largely neglected in previous phylogenetic studies.
INTRODUCTION
The Balkan Peninsula is a hotspot of genetic and species diversity and was a significant source of lineage dispersals to northerly adjacent areas after the end of the Pleistocene glaciations (Taberlet & al., 1998; Willner & al., 2009; Hewitt, 2011; Nieto Feliner, 2014; Španiel & Rešetnik, 2022). It is undoubtedly one of Europe's floristically richest regions, harbouring many endemic species (Kryštufek & Reed, 2004; Stevanović & al., 2007; Hewitt, 2011; Frajman & al., 2014). Several phylogenetic studies in the last decades revealed complex phylogeographic patterns within the Balkan Peninsula and indicated increased genetic diversity in the southern compared to the genetically more uniform northern areas (Caković & al., 2015; Đurović & al., 2017, 2021; Niketić & al., 2022; Španiel & Rešetnik, 2022). However, most of these studies were dedicated to plants distributed in the western and central parts of the peninsula, whereas the southern and eastern areas remained neglected in phylogenetic studies (Španiel & Rešetnik, 2022). The few existing studies including populations from the central, eastern, and southern Balkan Peninsula showed a clear differentiation between the western, mostly Dinaric populations and more eastern or southern Balkan populations, such as in the Sesleria rigida Heuff. complex (Poaceae; Kuzmanović & al., 2013), in Aurinia saxatilis (L.) Desv. (Brassicaceae; Rešetnik & al., 2022) or Cerastium decalvans Schloss. & Vuk. s.l. (Caryophyllaceae; Niketić & al., 2022). Cryptic diversity in the Cyanus tuberosus (Vis.) Soják group (Asteraceae), where groups of populations in the central and eastern Balkan Peninsula form distinct, mostly allopatric lineages, led to recognition of several species (Skokanová & al., 2019). Existing studies, although scarce, thus uncovered deep phylogenetic splits within the south-eastern Balkan Peninsula.
The eastern parts of the Balkans are peculiar due to the existence of extensive lowlands, compared to the other, highly mountainous regions. The Danubian and Thracian lowlands in Bulgaria and Romania are the south-westernmost extensions of the Eurasian zonal steppes, which are the second-largest continuous biome on Earth, covering up to 7% of the Earth's total land surface (Wesche & al., 2016). The steppes are extraordinarily species-rich habitats, known to harbour up to 72 herbaceous plant taxa per square metre, thus surpassing even the diversity of tropical rainforests at the comparable spatial scale (Wilson & al., 2012). They fulfil significant ecological functions, such as water regulation, carbon sequestration, and soil stability (Vorontzova & Zaugolnova, 1985; Kirschner & al., 2020; Török & al., 2020). With the exception of the easternmost Balkan Peninsula (Dobrudja), which is considered part of the macroclimatically determined Eurasian zonal steppes, extra-zonal steppe outposts embedded in a matrix of forest vegetation are disjunctly distributed in the eastern and southern parts of the Balkans (Kirschner & al., 2020). Recent phylogenetic studies of steppe plant species from this area showed deep divergence between (1) populations from the zonal steppes plus the Pannonian steppes (Euphorbia seguieriana Neck.; Euphorbiaceae) and populations from grasslands of the central, eastern and southern Balkan Peninsula (Euphorbia niciciana Borbás; Frajman & al., 2019), (2) the central Balkan plus the Pannonian populations and the zonal steppe populations in Euphorbia glareosa Pall. s.l. (Stojilkovič & al., 2022), and (3) the central Balkan populations and the zonal steppe plus the Pannonian populations in Astragalus onobrychis L. (Záveská & al., 2019). Altogether, this indicates a peculiarity of the central Balkan area, but also its incongruent phylogeographic position in relation to more eastern and northern steppes.
One of the plant groups exhibiting high species diversity in the Eurasian steppes is Euphorbia L. sect. Esula (Pers.) Dumort. (Riina & al., 2013; Heimer & Frajman, 2023). With about 100 species, this section is the second-largest within E. subg. Esula Pers. (Riina & al., 2013) and it is characterised by an exceptionally high incidence of polyploidy (Heimer & Frajman, 2023). Polyploidisation is one of the major evolutionary drivers of angiosperm diversification, and whole-genome duplications (WGDs) have occurred in all extant angiosperm lineages (Soltis & al., 2009; Wood & al., 2009; Wendel, 2015). By establishing immediate reproductive barriers, polyploidy provides opportunities for rapid speciation and has been involved in approximately 15% of speciation events in angiosperms (Ramsey & Schemske, 1998; Wood & al., 2009). The evolutionary consequences of polyploidisation are diverse and can act at genetic, morphological and ecological levels (Balao & al., 2011; Weiss-Schneeweiss & al., 2013). Thus, polyploids can exploit new niches, often have wider distributions compared to their diploid ancestors, and are more likely to become invasive (Hijmans & al., 2007; Leitch & Leitch, 2008; Van de Peer & al., 2017; Cheng & al., 2021; Heimer & Frajman, 2023).
In Euphorbia sect. Esula, extensive polyploidisations have been evidenced especially in the Eurasian Clade, where besides diploids (2n = 2x = 20), several tetra- (2n = 4x = 36, 40) and hexaploid (2n = 6x = 60) taxa or populations within heteroploid species have been found (Rice & al., 2015 and references therein; Heimer & Frajman, 2023). Species of the Eurasian Clade mostly grow in grasslands, but can also be found in open forests, semi-deserts, as well as riparian vegetation (Riina & al., 2013). Several species, especially those including higher ploidies, have extensive ranges, such as E. cyparissias L., which extends across most of Europe (Meusel & al., 1978; Pungaršek & Frajman, 2024), and E. virgata Waldst. & Kit., which is widespread in the steppes of Eurasia (Govaerts & al., 2000; Geltman, 2020), and has become a noxious and invasive weed in North America (Berry & al., 2016).
One of the heteroploid species from Euphorbia sect. Esula that includes di- and tetraploid populations (2n = 4x = 40; Stahevitch & al., 1988) is E. agraria M.Bieb., which is a typical species of the westernmost zonal steppes (Ukraine, Romania), but extends its range also to the extra-zonal steppes and grasslands of the eastern and southern Balkan Peninsula (Albania, Bulgaria, Greece, North Macedonia, Romania, Turkey) and westernmost Asia Minor (Turkey; Prokhanov, 1949; Radcliffe-Smith & Tutin, 1968; Radcliffe-Smith, 1982; Micevski, 1998; Strid, 2024). It occurs as an alien species also in North America (Berry & al., 2016). Due to high morphological variability, the populations of E. agraria from the central Balkan Peninsula (Kosovo, Montenegro, North Macedonia, Serbia) have been segregated as E. agraria var. subhastata (Vis. & Pančić) Griseb. (Radcliffe-Smith & Tutin, 1968; Janković & Nikolić, 1972; Matevski & Teofilovski, 2004; Euro+Med Plantbase, 2006–; Caković & Stešević, 2021). This variety, which mainly grows in open forests and stabilised screes as well as in ruderal places, is suggested to be distinguished from the typical variety by having lingulate-panduriform leaves, whereas the typical variety has triangular-ovate to oblong leaves (Radcliffe-Smith & Tutin, 1968; Stevanović & al., 2014). In addition, E. agraria var. thyrsiflora (Griseb.) Hayek with more elliptic leaves has been recognised in Bulgaria (Kuzmanov, 1979). Finally, E. agraria var. euboea (Halácsy) Hayek was based on a collection of specimens having linear-lanceolate leaves (Radcliffe-Smith & Tutin, 1968) from Steni at the western foot of Mt Dirfis on the island of Evvia (Euboea) in Greece by Pichler. However, since E. agraria was never found on Evvia again, it is likely that the specimens were collected by Pichler in the Athos Peninsula in 1873; “In some cases he [Pichler] is known to have mixed up localities and labels, so there is a slight possibility that the Euphorbia was in fact collected in Athos” (A. Strid, e-mail communication to B. Frajman on 30 June 2023).
Little is known about how morphological variability is related to the distribution of Euphorbia agraria and E. subhastata, and to ploidy variation. A tetraploid chromosome number of 2n = 4x = 40 has been reported for E. agraria (Stahevitch & al., 1988), but Heimer & Frajman (2023), using relative genome size estimation, have shown that some diploid populations exist in the Balkan Peninsula in addition to the more common tetraploid ones. Based on our field observations, the morphological differentiation between the western and eastern populations in the Balkans is also accompanied by ecological differentiation, since in the western parts of the range the species primarily grows on stabilised screes and in open thermophilous forests in river gorges of the Dinaric Mountains (Bosnia, Montenegro, Serbia, Kosovo, Albania and North Macedonia), whereas in the east it mostly thrives in grasslands (authors’ personal observations; see also Stevanović & al., 2014). The western populations might thus be phylogenetically divergent and merit recognition as a distinct species, E. subhastata, as originally treated by Visiani & Pančić (1862) and suggested by Stevanović & al. (2014). The phylogenetic relationships between both taxa based on internal transcribed spacer (ITS) sequences and a few studied populations, as well as their exact phylogenetic position within the Eurasian Clade remained unresolved (Heimer & Frajman, 2023).
Considering the uncertain evolutionary origin and differentiation of Euphorbia agraria s.l. as well as the heterogenous phylogeographic patterns inferred for other grassland species in the central and eastern Balkan Peninsula (Frajman & al., 2019; Záveská & al., 2019; Stojilkovič & al., 2022), we here apply an integrative approach to disentangle the relationships among the populations sampled across this area. We (1) first estimated the ploidy of all investigated populations via relative genome size (RGS) estimation with flow cytometry. (2) Using nuclear ITS sequences and amplified fragment length polymorphism (AFLP) fingerprinting we inferred the origin and phylogeographic differentiation within the study species. Finally (3) using multivariate morphometrics we explored the morphological differentiation and (4) based on all data propose a revised taxonomic treatment.
MATERIALS AND METHODS
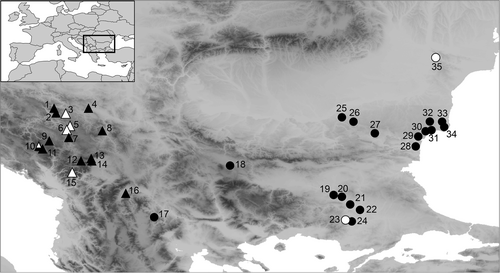
Plant material
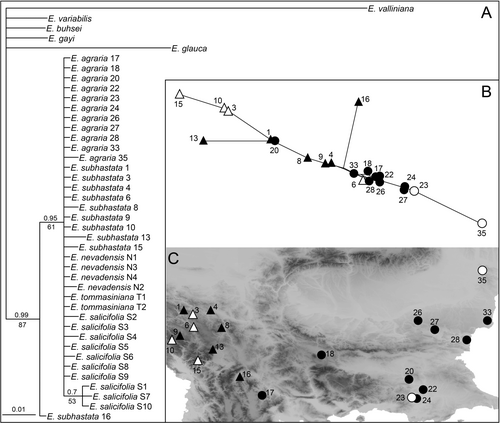
We collected the plant material in the field between 2013 and 2023. For molecular and RGS analyses, we dried the leaves in silica gel, whereas for the morphometric measurements we prepared herbarium vouchers. We sampled 35 populations of Euphorbia agraria s.l.; all were included in the RGS and AFLP analyses, and 22 in the morphometric analyses (Fig. 1, suppl. Table S1, Appendix 1). We identified the plants using the diagnostic characters outlined above (Radcliffe-Smith & Tutin, 1968; Janković & Nikolić, 1972; Kuzmanov, 1979; Stevanović & al., 2014), and for simplicity and in anticipation of our results we refer to them as E. agraria and E. subhastata hereafter. We sampled E. subhastata across its entire distribution, including the locus classicus (population 4), whereas for E. agraria the sampling was focused on its westernmost distribution range. We thus sampled the only known population (17) from North Macedonia (Micevski, 1998), the entire distribution range in Greece (Strid, 2024), as well as one of the westernmost populations in Bulgaria (18) based on the review of specimens in the herbarium of the Bulgarian Academy of Sciences (SOM) and the notion in the Bulgarian Flora (Kuzmanov, 1979) that the species is absent from the western and southern mountainous areas. In the Danubian and Thracian lowlands the species is more widespread (Kuzmanov, 1979), but we sampled only several populations from both areas as our aim was to explore its divergence from E. subhastata, rather than infraspecific differentiation within E. agraria. Finally, we included E. tommasiniana Bertol., E. nevadensis Boiss. & Reut. and E. salicifolia Host. from E. sect. Esula, as most closely related outgroups in the RGS, AFLP and ITS analyses (suppl. Table S1), based on preliminary results of RAD sequencing data (B. Frajman, unpub.).
Relative genome size and ploidy level estimation
We measured RGS using a CyFlow space flow cytometer (Partec, Münster, Germany) following Suda & Trávníček (2006). Nuclei from silica gel-dried leaf material of Euphorbia agraria (19 populations) and E. subhastata (15; suppl. Table S1) and fresh leaves of the reference standard Bellis perennis L. (2C = 3.38 pg; Schönswetter & al., 2007) were stained using 4′,6-diamidino-2-phenylindole (DAPI). For comparison, we also measured RGS of closely related E. nevadensis (5 populations), E. salicifolia (10) and E. tommasiniana (2). We recorded the relative fluorescence of 3000 nuclei for three to five individuals per population and used FloMax software (Partec) to evaluate histograms and calculate coefficients of variation. The RGS was calculated as the ratio between the values of the mean relative fluorescence of the sample and the standard. For statistical analyses of RGS data we used RStudio v.1.2.5019 (RStudio Team, 2022), with the visualization package ggplot2 to produce scatter and box plots. We checked diploids and tetraploids separately for normality using a Shapiro-Wilk normality test, for homogeneity of variances using a Levene test, and for the significance of differences using a T test.
DNA extraction, ITS sequencing, and analyses of sequence data
Extraction of total genomic DNA and ITS sequencing were performed as described by Frajman & Schönswetter (2011), with the exception that sequencing was carried out at Eurofins Genomics (Ebersberg, Germany). Contigs were assembled, edited, and sequences aligned using Geneious Pro v.5.5.9 (Kearse & al., 2012). Base polymorphisms were coded using NC-IUPAC ambiguity codes. We produced 7 ITS sequences of E. agraria, 6 of E. subhastata, 2 of closely related E. nevadensis and 1 of E. salicifolia. We added 4 sequences of E. agraria, 4 of E. subhastata, and 18 of other taxa from E. sect. Esula from Heimer & Frajman (2023; GenBank numbers in Appendix 1 and suppl. Table S1). As relationships within the Eurasian Clade including E. agraria s.l. were largely unresolved in the study of Heimer & Frajman (2023), we here only included E. nevadensis, E. salicifolia and E. tommasiniana that are closely related to E. agraria s.l. based on RAD sequencing results (Frajman, unpub.). In addition, more distantly related E. buhsei Boiss., E. gayi Salis, E. valliniana Belli and E. variabilis Ces. were used for rooting. Maximum parsimony (MP) and maximum parsimony bootstrap (MPB) analyses were performed using PAUP v.4.0b10 (Swofford, 2002) as described by Frajman & al. (2019). Bayesian analyses were performed using MrBayes v.3.2.1 (Ronquist & al., 2012) applying the HKYI substitution model and the settings as in Frajman & al. (2019). We also produced a NeighbourNet with ITS sequences of E. agraria s.l. using SplitsTree4 v.12.3 (Huson & Bryant, 2006).
AFLP analyses
The AFLP procedure followed Vos & al. (1995) with the modifications described by Cresti & al. (2019). The three primers for selective PCR (fluorescent dye in parenthesis) were EcoRI (Fam6)-ATC/MseI-CTC, EcoRI (Vic1)-ACG/MseI-CAA, and EcoRI (Ned2)-ACC/MseI-CAT. Subsequently, 1.3 μl of the elution product was mixed with 10 μl formamide and 0.13 μl GeneScan 500 ROX (ThermoFisher Scientific, Waltham, Massachusetts, U.S.A.) and run on a 3130xl Genetic Analyzer (Applied Biosystems, Waltham, Massachusetts, U.S.A.). One blank (DNA replaced by water) was included to test for contamination and 17 samples were used as replicates between the two PCR batches to test the reproducibility. Electropherograms were analysed with Peak Scanner 1.0 (Applied Biosystems) using default peak detection parameters except for employing light peak smoothing. Automated binning and scoring of the AFLP fragments were performed using RawGeno v.2.0-1 (Arrigo & al., 2009) for RStudio v.2022.12.0+353 (RStudio Team, 2022) with the following settings: scoring range 75–500 bp, minimum intensity 100 RFUs, minimum bin width 1 bp and maximum bin width 1.5 bp. Fragments with a reproducibility less than 80% based on sample-replicate comparisons were eliminated. The error rate (Bonin & al., 2004) was calculated as the ratio of mismatches (scoring 1 vs. 0) over phenotypic comparisons in AFLP profiles of replicated individuals. A matrix of 185 individuals was finally produced and analysed as described below. A neighbour-joining (NJ) analysis based on Nei-Li genetic distances (Nei & Li, 1979) was conducted and bootstrapped (2000 pseudo-replicates) with TREECON v.1.3b (Van de Peer & De Wachter, 1997), using E. nevadensis, E. salicifolia and E. tommasiniana for rooting. SplitsTree4 v.12.3 (Huson & Bryant, 2006) was used to produce NeighbourNets based on uncorrected P-distances separately for E. agraria and E. subhastata that were clearly monophyletic and divergent based on the NJ analysis (see Results). For the same reason, we also performed non-hierarchical K-means clustering (Hartigan & Wong, 1979) for both taxa separately, using a script of Arrigo & al. (2010) in RStudio v.2022.12.0+353 (RStudio Team, 2022). A total of 50,000 independent runs were performed (i.e., starting from random points) for each assumed value for K clusters ranging from 2 to 10. To select the best number of groups, the strategy proposed by Evanno & al. (2005) was used and the proportions of individuals assigned to K-means groups (within populations) were displayed on a map in ArcMAP v.10.8.2 (https://desktop.arcgis.com).
Morphometric analyses
We performed morphometric analyses on 22 individuals from 12 populations of Euphorbia agraria and 18 individuals from 10 populations of E. subhastata, thus including one to two (in one population four) individuals per population in the analyses. In total, we measured 36 characters and calculated 18 ratios (suppl. Table S2). Manual measurements were done for stem and leaf characters, while cyathium, fruit and seed characters were measured from images taken with an Olympus SZX9 stereomicroscope (Olympus, Hamburg, Germany) and analysed with the Olympus image analysis software analySIS pro v.3.2. In E. subhastata, in two individuals from population 13, cyathium, fruit and seed characters were not developed, and in one individual from population 9, fruits and seeds were not developed. We replaced these missing values with the means of other individuals of E. subhastata.
Statistical analyses were performed using SPSS v.26.0 (https://www.ibm.com/de-de/products/spss-statistics). Correlation between the metric traits was assessed using Pearson and Spearman correlation coefficients, with six pairs of characters showing a correlation exceeding 0.82 in one or both correlation coefficients. The following characters were therefore excluded from further analyses: Stem length, Depth of the basal emargination (length of auricles), Distance from the base to the widest part of a raylet leaf and Caruncle length. After standardization to a mean of zero and a variance of one unit, we performed principal component (PCA) and discriminant analysis (DA). Based on the morphometric data, we produced taxon descriptions and an identification key. The metric values shown correspond to the 10th and 90th percentiles, supplemented by extreme values in parentheses.
RESULTS
Relative genome size variation
The RGS values were discretely distributed among the studied populations (suppl. Table S1; Fig. 2). In diploid populations of Euphorbia agraria RGS ranged from 0.404 to 0.411, and in tetraploids from 0.797 to 0.857. In diploid E. subhastata RGS ranged from 0.435 to 0.445, and in tetraploids from 0.785 to 0.889. In outgroup species, RGS of three diploid populations of E. nevadensis was 0.408–0.414, one population was putatively triploid with RGS 0.615, and one tetraploid with RGS 0.809. In four diploid populations of E. salicifolia RGS was 0.429–0.442, whereas six tetraploid populations had RGS 0.827–0.885. Two diploid populations of E. tommasiniana had RGS 0.401 and 0.413.
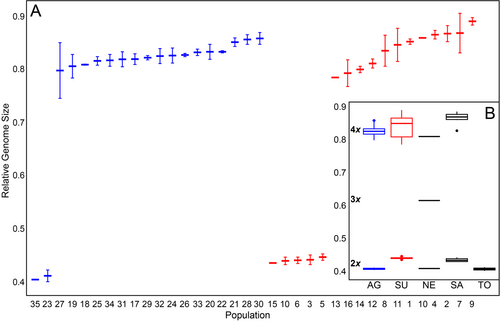
The RGS of diploid Euphorbia agraria and E. subhastata was not normally distributed (Shapiro-Wilk test: p = 0.04515) and their variance was homogeneous (Levene test: p = 0.1681). Furthermore, the difference in diploid RGS between the two taxa was not significant (T-test: p = 0.0892). The RGS of tetraploid populations within both taxa was normally distributed (Shapiro-Wilk test: p = 0.3828) and their variance was homogeneous (Levene test: p = 0.7939). In addition, the difference in RGS between tetraploid populations of both taxa was not significant (T-test: p = 0.2467).
Phylogenetic relationships based on AFLPs
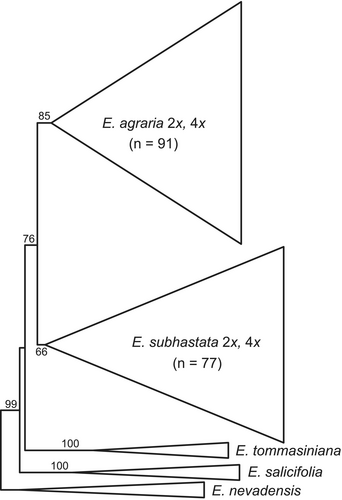
A total of 525 fragments were scored in 186 individuals; 48 fragments were excluded because they were present or absent in a single individual only. The error rate (Bonin & al., 2004), calculated before the exclusion of non-reproducible fragments and based on 17 replicates, was 3.7%. In the NJ tree (Fig. 3; suppl. Fig. S1) both Euphorbia agraria and E. subhastata were monophyletic (bootstrap support, BS, 85% and 66%, respectively). Together, they formed a sister cluster (BS 76%), with E. salicifolia (BS 100%) and E. tommasiniana (BS 100%) being more closely related (BS 99%), whereas E. nevadensis was more distant. Within E. agraria and E. subhastata, most of the clusters had low bootstrap support, with the exception of some terminal clusters mostly including individuals of the same populations that had moderate to high support. In E. agraria the two diploid populations were positioned close in the tree, mostly together with geographically close tetraploid populations, whereas in E. subhastata there were two distant groups of diploid populations nested within tetraploid populations; in both cases the support for these clusters was low (BS < 50%).
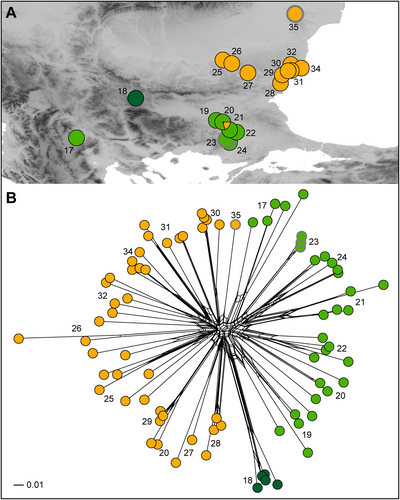
Non-hierarchical K-means clustering revealed an optimal separation into three groups for Euphorbia agraria (Fig. 4A), whereas four or more groups had much lower delta K. One cluster (dark green in Fig. 4A) included a single population from western Bulgaria. Another cluster (light green) included one population from North Macedonia, four populations from Greece (Thrace region; including one diploid population), and two populations from adjacent Bulgaria; one of the latter populations was admixed with the third cluster. The third cluster (orange) was composed of ten tetraploid populations from eastern Bulgaria and one diploid population from Romania. The three clusters were reflected also in the structure revealed in the NeighbourNet (Fig. 4B). In E. subhastata the optimal division was into two clusters (Fig. 5A). One (red in Fig. 5A) included three tetraploid populations, two from Kosovo and one from North Macedonia. The other cluster (blue) included the other tetraploid populations and all diploid populations. The clustering was less clearly reflected in the NeighbourNet (Fig. 5B), where population 11 from the blue cluster was closely related to populations 12 and 14 from the red cluster, whereas population 16 from North Macedonia was more divergent. In general, the NeighbourNet of E. subhastata indicated a more pronounced differentiation among (groups of) populations compared to E. agraria.
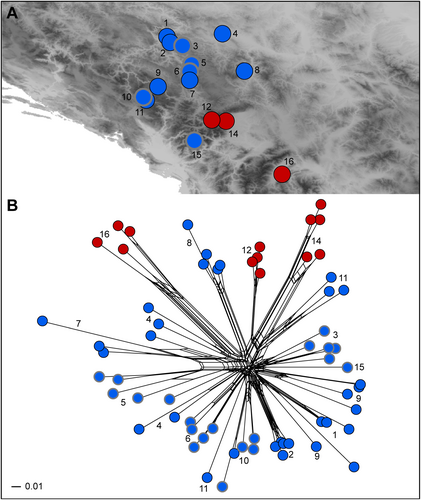
Phylogenetic relationships based on ITS
The ITS alignment (suppl. Appendix S1) was 704 bp long. The trees inferred by parsimony and Bayesian analyses were largely congruent (Fig. 6A), but the relationships were poorly resolved and all accessions of E. agraria and E. subhastata, with exception of E. subhastata 16, were in a polytomy (posterior probabilities, PP 0.95, BS 61%) with all accessions of E. nevadensis, E. salicifolia and E. tommasiniana. The geographically most isolated accession 16 of E. subhastata from North Macedonia was sister to the polytomic clade (PP 0.99, BS 87%). The divergent position of population 16 was based on a G at position 115 in the alignment, which was shared with E. gayi and other species used for rooting, whereas all other accessions had either T (most accessions) or K (polymorphism for T/G present in E. agraria 18 and E. salicifolia S9) at this position. Euphorbia subhastata 16 was also divergent in the NeighbourNet. All other accessions were distributed along the roughly linear network, in which most accessions of E. subhastata were positioned on one side and most accessions of E. agraria on the other side of the network, with diploids of each species being most divergent (with exception of population 6). Exceptions, besides population 16, were population 20 of E. agraria, which was positioned at the branch including E. subhastata, and the diploid accession 6 of E. subhastata, which was positioned among most samples of E. agraria.
Morphological differentiation
The character states for morphological characters, including ratios, are presented in suppl. Table S2. Most individuals of Euphorbia agraria and E. subhastata were separated in the PCA scatter plot (Fig. 7A) along the first component, but not along the second and third; the first three components explained 19.8%, 13.5% and 10.7% of the total variation. The characters contributing most to this separation were: Ratio Length of the cyathial involucre/Width of the cyathial involucre, Ratio Distance from the base to the widest part of a ray leaf/Length of a ray leaf, Fruit length, Fruit width, Seed length, Seed width, Length of cyathial gland without appendage (horn), Width of cyathial gland, Depth of the basal emargination of the stem leaf (length of auricles), Caruncle width, Number of sterile axillary rays.
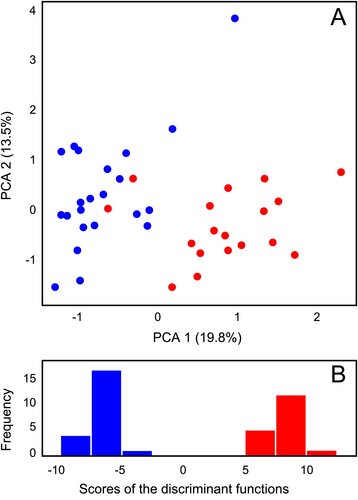
The DA (Fig. 7B) showed a clear morphological divergence between the two species. The characters contributing most to this divergence were: Distance from the base to the widest part of a middle stem leaf, Length of a middle stem leaf, Ratio of Distance from the base to the widest part of a middle stem leaf/Length of a middle stem leaf, Ratio of Distance from the base to the widest part of a ray leaf/Length of a ray leaf, Fruit width, Length of cyathial gland without appendage (horn), and Distance from the base to the widest part of a ray leaf.
Finally, also BoxPlots (suppl. Fig. S2) showed that the following characters are divergent between both taxa: Ratio Length of cyathial involucre/Width of cyathial involucre, Ratio of Distance from the base to the widest part of a ray leaf/Length of a ray leaf, Fruit length, Seed length, Seed width, Length of cyathial gland without appendage (horn), Width of cyathial gland, Depth of the basal emargination (length of auricles), Number of sterile axillary rays and Caruncle width.
DISCUSSION
Our integrative approach including phylogenetic, cytogenetic, and morphometric analyses revealed a clear differentiation between Euphorbia subhastata endemic to the central Balkan Peninsula, and more widespread south-eastern and eastern European E. agraria. Our phylogenetic analyses of the AFLP data (Fig. 3, suppl. Fig. S1) indicated that E. tommasiniana from the north-westernmost Balkan Peninsula and E. salicifolia from the central Balkan Peninsula are their next relatives. In addition, AFLP data indicated a clear divergence between both taxa, correlated with a narrow distribution gap between them in North Macedonia, and a much broader gap in Serbia (Fig. 1) (Stevanović & al., 2014). This is in line with the few existing studies focusing on the area inhabited by E. agraria and E. subhastata, which showed a clear differentiation in different plant species groups between the western, mostly Dinaric populations, and the more eastern or southern Balkan populations (Kuzmanović & al., 2013; Záveská & al., 2019; Niketić & al., 2022; Rešetnik & al., 2022). These results, along with those presented here, highlight the peculiarity of the central Balkan Peninsula in relation to more extensive steppe areas of the eastern Balkan Peninsula and beyond.
The geographic complexity of the central Balkan Peninsula, which is characterised by a heterogeneous mosaic of mountain ridges and deep river gorges (Turrill, 1929; Horvat & al., 1974; Frajman & Oxelman, 2007; Caković & al., 2015), is reflected also in the pronounced genetic differentiation within Euphorbia subhastata (Fig. 5B). On the other hand, the differentiation among populations of E. agraria, with the exception of the disjunct western populations 17 and 18, is less pronounced (Fig. 4B). Existence of extensive lowlands such as the Danubian and Thracian lowlands in the eastern Balkans enables better connectivity and facilitates gene flow among populations of E. agraria and accompanying grassland species. Nevertheless, the K-means analysis revealed genetic differentiation of the more northern Danubian populations 25–35 from the more southern Thracian populations 19–24 (Fig. 4), which might be a result of differential Pleistocene refugia south and north of Stara Planina mountain range in central Bulgaria.
Genetic divergence accompanied by ecological differentiation into screes, open forests and scrublands of river gorges in Euphorbia subhastata vs. grasslands and partly ruderal paces such as road margins and abandoned fields in E. agraria, triggered also morphological differentiation between both species (Fig. 7). Our DA (Fig. 7B) indicated that the leaf shape is one of the most important characters differentiating both species. Whereas the leaves of E. subhastata are mostly longer and lingulate-panduriform, those of E. agraria are shorter and mostly triangular-ovate. On the other hand, the difference in the branching patterns indicated by Stevanović & al. (2014) is not as consistent, as there are also individuals of E. agraria that bear non-flowering axillary shoots, along the flowering axillary rays. Nevertheless, E. subhastata is usually more richly branched with a higher number of vegetative axillary shoots. Finally, also fruit and seed dimensions were shown to partly discriminate between the two species in the PCA and BoxPlots (suppl. Fig. S2). On the other hand, the species do not differ in their RGS, neither at diploid nor at the tetraploid level; their RGS is comparable with RGSs of closely related E. nevadensis, E. salicifolia and E. tommasiniana (Fig. 2).
The existence of diploid and tetraploid populations in both species indicates independent polyploidisations within each of them. The fact that different diploid populations are intermingled with tetraploid ones in the AFLP NeighbourNets (Figs. 4, 5) suggests multiple autotetraploidisation events within each species. Such a pattern was also observed in Euphorbia montenegrina (Bald.) K.Malý from the Balkan Peninsula (Caković & al., 2021) and suggested for several species of E. sect. Esula, where occurrence of multiple ploidy levels within single species is a common phenomenon and recurrent polyploidisations are rather a rule than an exception (Heimer & Frajman, 2023; Pungaršek & Frajman, 2024).
In summary, based on our integrative approach, we show that Euphorbia agraria and E. subhastata are distinct allopatric evolutionary lineages, which are morphologically and ecologically divergent. Therefore, we propose to treat E. subhastata as a distinct species, as originally described by Visiani & Pančić (1862) and later suggested by Stevanović & al. (2014). Our results thus support the view that the Balkan Peninsula is a hotspot of genetic and species diversity and indicate that future biodiversity research of this part of Europe should focus on the areas of the central, eastern and southern Balkan Peninsula that were largely neglected in previous phylogenetic studies focussing on the Dinaric Mountains (Španiel & Rešetnik, 2022).
TAXONOMIC TREATMENT
Identification key
1. Stems (2.1)2.6–5.7(6.5) mm thick, with 0–2(3) axillary vegetative shoots, which are (0.7)2.8–8.4(9.0) cm long, and (0)2–22(25) fertile axillary rays. Terminal rays (1.7)2.6–12.4(15) cm long. Cauline leaves triangular-ovate to oblong, rarely linear-lanceolate, 1.5–2.8(3.3) times longer than wide. Cyathial glands (0.4)0.5–2.0(2.3) × (0.9)1.0–3.0(3.4) mm E. agraria
Euphorbia agraria M.Bieb., Fl. Taur.-Caucas. 1: 375. 1808 ≡ Tithymalus agrarius (M.Bieb.) Klotzsch & Garcke in Abh. Königl. Akad. Wiss. Berlin 1859(1): 89. 1860 – Lectotype (designated by Geltman in Novosti Sist. Vyssh. Rast. [Novit. Syst. Pl. Vasc.] 51: 68. 2020): “Ex Tauria, 1794 [Marschall Bieberstein]” (LE barcode LE 01070980 n.v.).
= Euphorbia thyrsiflora Griseb., Spic. Fl. Rumel. 1: 143. 1843 ≡ E. agraria var. thyrsiflora (Griseb.) Nyman, Consp. Fl. Eur.: 653. 1881 – Lectotype (designated by Strid in Preslia 72: 288. 2000): “Ainadgik” [the rest of the text illegible], Grisebach no. 286 (GOET barcode GOET003702 [image!]).
= Euphorbia transsilvanica Schur in Verh. Mitth. Siebenbürg. Vereins Naturwiss. Hermannstadt 3: 124. 1852 – Lectotype (designated here): “[Romania] in collibus ad margines agrorum prope pagum Hammersdorf”, Jul, Schur s.n. (LW barcode LW00211309 [image!]).
= Euphorbia euboea Halácsy, Consp. Fl. Graec. 3: 107. 1904 – Lectotype (designated here): “Griechenland. Auf waldigen Orten bei Stenni”, Jun 1876, Th. Pichler s.n. (WU No. 0078183 [image!]).
Additional material examined
Euphorbia transsilvanica Schur, “[Romania] in collibus argillensis prope pagum Hammersdorf pusta Bibiniam”, Jul, Schur s.n. (LW barcode LW00211308 [image!]).
Notes
Population 19 from Bulgaria, which corresponds morphologically to the description of Euphorbia thyrsiflora, is phylogenetically nested among other samples. Given the species’ morphological variability and the local occurrence of different morphotypes it is best to treat E. agraria as a species without infraspecific taxa. In addition, “Euphorbia nitens Trevir. in Mag. Neuesten Entdeck. Gesammten Naturk. Ges. Naturf. Freunde Berlin 7: 149 (1816)” is listed in synonymy of E. agraria at POWO (2024). However, “fol[ius]. […] lucidis, coriaceis” and the fact that the author compares this species with E. nicaeensis in the protologue and lists Caucasus as the occurrence area (where E. agraria does not occur; see Geltman, 2020), suggest that this is rather a synonym of E. glareosa Pall. ex M.Bieb.
Description
Glabrous perennial, (33)34–67(73) cm high, stems erect, (24)30–61(62) cm long and (2.1)2.6–5.7(6.5) mm thick. Stem with 0–2(3) axillary vegetative shoots, (0.7)2.8–8.4(9.0) cm long, and (0)2–22(25) fertile axillary rays, (2.2)4.0–13.8(28.0) cm long. Lowest axillary rays at (12)16.2–41.7(47.4) cm from the base of the stem, i.e., at (0.3)0.4–0.9(1.0) of the stem length. Terminal rays (3)4–14(17), 0–2 times dichotomously branched, (1.7)2.6–12.4(15) cm long, (1.9)2.6–8.2(10.2) times longer than ray leaves. Cauline leaves (18)22–47(49) × (7)9.5–22.5(31) mm, triangular-ovate to oblong, rarely linear-lanceolate, 1.5–2.8(3.3) times longer than wide, widest at 0.2–0.6 of their length. Leaf base broadly cordate-auriculate, amplexicaul, basal emargination (0.3)0.7–3.2(3.7) mm deep. Ray leaves triangular or ovate, broadly cordate, (7.9)8.2–19.3(27) × (4.3)6.2–12.0(16.2) mm, (0.7)0.9–2.0(2.2) times longer than wide, widest at 0.1–0.5(0.6) of their length. Raylet leaves triangular-subreniform, 6.0–9.4(11.0) × (8.1)9.7–18.5(19.3) mm, 0.35–0.75 times longer than wide, widest at (0.1)0.2–0.6(0.8) of their length. Cyathial involucre (1.9)2.2–5.9(7.1) × (1.2)1.5–3.8(6.1) mm, (0.6)0.9–2.5(3.3) times longer than wide. Cyathial glands obovate-truncate, (0.4)0.5–2.0(2.3) × (0.9)1.0–3.0(3.4) mm, (0.9)1.1–4.2(5.3) times broader than long, with (0.4)0.5–1.0(1.1) mm deep emargination. Fruits glabrous, granulate, broadly ovate, 2.3–3.7(4.2) × 1.5–3.2(5.0) mm, (0.7)0.8–2.0(2.1) times longer than wide, widest at (0.2)0.3–0.5(0.6) of the length. Styles (1.4)1.5–3.1(5.0) mm long. Seeds ovoid, smooth, grey, 2.1–2.8(3.4) × 1.4–1.9(2.4) mm, 1.3–1.8 times longer than wide, widest at (0.2)0.3–0.5 of the length. Caruncle conical, 0.4–0.6(0.7) × 0.7–1.0 mm, (0.4)0.5–0.9 times longer than wide, widest at 0.2–0.5(0.7) of the length.
Distribution
Native to Bulgaria, Greece, Moldova, North Macedonia, Romania, Turkey, Ukraine (incl. Crimea) and possibly Russia (see Geltman, 2011). Introduced to several other countries (see Geltman, 2006, and POWO, 2024).
Habitat
Euphorbia subhastata Vis. & Pančić in Mem. Reale Ist. Veneto Sci. 10: 444, t. 7. 1861 ≡ Euphorbia agraria var. subhastata (Vis. & Pančić) Griseb. ex Asch. & Kanitz, Cat. Cormophyt. Anthophyt. Serbiae: 92. 1877 – Lectotype (designated by Clementi & al. in Phytotaxa 202: 130. 2015): “De rupib[us] M. Kablar Serb[ia] centr[alis] fl. culta”, s.d., J. Pančić s.n. (PAD barcode H0023182 [image!]).
Additional material examined
Euphorbia subhastata, [SERBIA]. s.l., s.d., s.c. s.n. (PAD H0023183!); SERBIA. M. Kablar, Jul [1]858, J. Pančić s.n. (BEOU No. 764!).
Description
Glabrous perennial, (24)32–60(67) cm high, stems erect, (18)28–52(59) cm long and (2.2)2.4–4.9 mm thick. Stem with 1–7(8) axillary vegetative shoots, (3.0)3.1–18.4(18.8) cm long, and (0)2–13(18) fertile axillary rays, (3.1)4.2–10.8(13) cm long. Lowest axillary rays at (10.8)11.8–39.2(40.7) cm from the base of the stem, i.e., at (0.3)0.4–0.9 of the stem length. Terminal rays (3)4–11(17), 0–2 times dichotomously branched, (3.4)3.5–8.1(8.3) cm long, (2.0)2.2–6.1(6.2) times longer than ray leaves. Cauline leaves (20)25–48(68) × (7.3)8.8–24(25) mm, lingulate-panduriform, rarely oblong, 1.6–4.3(6.0) times longer than wide, widest at (0.3)0.4–0.7(0.8) of their length. Leaf base cordate-auriculate, amplexicaul, basal emargination 0.5–3.2(3.8) mm deep. Ray leaves ovate to obovate, cordate, (8.8)9.1–21.0(21.4) × (3.8)4.0–10.1(10.7) mm, 1.1–3.1(4.4) times longer than wide, widest at 0.2–0.6(0.9) of their length. Raylet leaves triangular-subreniform (7.4)7.7–12.8(14.1) × (8.1)10.9–18.9(20.1) mm, 0.4–0.8(1.0) times longer than wide, widest at 0.1–0.5(0.7) of their length. Cyathial involucre (3.5)3.6–5.6(5.9) × (1.7)1.8–2.9(3.1) mm, (1.5)1.6–2.4 (2.8) times longer than wide. Cyathial glands obovate-truncate, (0.4)0.5–0.9(1.0) × 1.2–2.8 mm, 1.2–5.6(6.0) times broader than long, with 0.4–0.8 mm deep emargination. Fruits glabrous, granulate, broadly ovate, (1.4)1.5–3.9(4.0) × (1.2)1.3–3.1(3.2) mm, (0.6)0.9–1.9(2) times longer than wide, widest at 0.3–0.6 of the length. Styles 1.4–3.1(3.4) mm long. Seeds ovoid, smooth, grey, 2.2–3.1 × 1.4–2.0(2.1) mm, 1.2–1.9 times longer than wide, widest at (0.3)0.4–0.5(0.6) of the length. Caruncle conical, (0.3)0.4–0.7(0.8) × 0.7–1.2 mm, 0.3–0.9 times longer than wide, widest at 0.2–0.5 of the length.
Distribution
Albania, Kosovo, Montenegro, North Macedonia, Serbia, Bosnia and Herzegovina.
Habitat
Calcareous screes and rocky sites, open forests and scrublands in river gorges.
AUTHOR CONTRIBUTIONS
BF designed and coordinated the study, collected plants, performed phylogenetic analyses and wrote parts of the paper. NB performed field work, lab work and analyses of RGS, AFLP and morphometric data and wrote parts of the paper. PS advised the AFLP analyses and improved the final version of the manuscript.
ACKNOWLEDGEMENTS
N. Berisha's stay in Innsbruck was supported by a HERAS+ Post-PhD Scholarship via the ÖAD (MPC-2022-05431). B. Frajman's fieldwork was partly supported by the Austrian Agency for International Cooperation in Education and Research (ÖAD-GmbH; grant RS 07/2022 to PS and BF). D. Geltman helped us with the literature, N. Kuzmanović, M. Niketić, S. Đurović, A. Strid and A. Ivanova helped with locality data, and M. Appelhans, J. Danihelka, T. Khmil, C. Pachschwöll and M. Puscas with tracing the types. We thank the curators of the Herbarium of the Institute of Biodiversity and Ecosystem Research, Bulgarian Academy of Sciences (SOM) in Sofia for providing type images, and all collectors listed in Appendix 1/suppl. Table S1 for help with the collection of samples. D. Pirkebner, M. Magauer, and M. Barfuss helped with laboratory work and N. Kuzmanović with K-means analyses. M. Magauer prepared some figure drafts.
Appendix 1: Taxon, ID (when applicable), origin, voucher information (herbarium code) and ITS GenBank accession numbers (when applicable) for samples included in the study. Accessions of the newly sequenced DNA sequences are followed by asterisk. The samples without GenBank numbers were used in AFLP, relative genome size (RGS) and/or morphometric analyses. See suppl. Table S1 for full version
Euphorbia agraria M.Bieb. 17, North Macedonia, N. Berisha 17857 (IB), PQ497644*; 18, Bulgaria, B. Frajman 17782 (IB), JN010022; 19, Bulgaria, N. Berisha 17856 (IB), –; 20, Bulgaria, B. Frajman 17781 (IB), PQ497642*; 21, Greece, B. Frajman 17780 (IB), –; 22, Greece, B. Frajman 17779 (IB), PQ497641*; 23, Greece, B. Frajman 17777 (IB), PQ497640*; 24, Greece, B. Frajman 17776 (IB), PQ497639*; 25, Bulgaria, S. Đurović, U. Buzurović, M. Rat 15696 (IB), –; 26, Bulgaria, A. Ivanova 17889 (IB), PQ497645*; 27, Bulgaria, S. Đurović, U. Buzurović, M. Rat 15691 (IB), ON511318; 28, Bulgaria, N. Berisha 17849 (IB), PQ497643*; 29, Bulgaria, N. Berisha 17852 (IB), –; 30, Bulgaria, N. Berisha 17855 (IB), –; 31, Bulgaria, N. Berisha 17854 (IB), –; 32, Bulgaria, N. Berisha 17850 (IB), –; 33, Bulgaria, E. Kabaš, S. Vukojičić 15208 (IB), ON511317; 34, Bulgaria, N. Berisha 17851 (IB), –; 35, Romania, B. Frajman & P. Schönswetter 14193 (IB), ON511316; Euphorbia buhsei Boiss., Iran, B. Frajman, M. Falch & Ch. Gilli 14307 (IB), ON511321; Euphorbia gayi Salis, Italy, B. Frajman 16799 (IB), ON511349; Euphorbia glauca G.Forst, New Zealand, B. Frajman 15268 (IB), ON511351; Euphorbia nevadensis Boiss. & Reut. N1, Spain, P. Carnicero & D. Baumgartner 17954 (IB), PQ497647*; N2, Spain, B. Frajman 17226 (IB), PQ497646*; N3, Spain, R. Riina, J. Molero & P.E. Berry 14668 (IB), ON511363; N4, Spain, G. Nieto Feliner & I. Álvarez 17092 (IB), ON511364; Spain, J. Peñas de Giles 17953 (IB), –; Spain, P. Carnicero & D. Baumgartner 17955 (IB), –; Euphorbia salicifolia Host S1, Bosnia and Herzegovina, B. Frajman 15094 (IB), ON511369; S2, North Macedonia, B. Frajman 17379 (IB), PQ497648*; S3, Austria, L. Schratt-Ehrendorfer 11107 (IB), JN010090; S4, Bosnia and Herzegovina, B. Frajman & P. Schönswetter 12394 (IB), JN010088; S5, Greece, B. Frajman 14767 (IB), JN010091; S6, Bulgaria, B. Frajman & P. Schönswetter 11305 (IB), JN010091; S7, Greece, B. Frajman 15657 (IB), ON511370; S8, Kosovo, P. Schönswetter, B. Frajman & D. Kutnjak 12963 (IB), ON511367; S9, Montenegro, M. Niketić, G. Tomović, M. Turjak & B. Frajman 11932 (IB), JN010089; S10, North Macedonia, B. Frajman 15659 (IB), ON511371; S11, Montenegro, B. Frajman & P. Schönswetter 14526 (IB), –; S12, Serbia, B. Frajman 14978 (IB), –; S13, Serbia, S. Đurović 15209 (IB), –; S14, Serbia, B. Frajman 17739 (IB), –; S15, Serbia, B. Frajman 17787 (IB), –; Euphorbia subhastata Vis. & Pančić 1, Serbia, B. Frajman 15782 (IB), ON511372; 2, Serbia, B. Frajman 17742 (IB), –; 3, Serbia, B. Frajman 17743 (IB), PQ497649*; 4, Serbia, B. Frajman 17753 (IB), PQ497651*; 5, Serbia, B. Frajman 17747 (IB), –; 6, Serbia, B. Frajman 17748 (IB), PQ497650*; 7, Serbia, B. Frajman 17751 (IB), –; 8, Serbia, S. Đurović, S. Vukojičić 15831 (IB), ON511373; 9, Montenegro, D. Caković 17916 (IB), PQ497654*; 10, Montenegro, B. Frajman 17905 (IB), PQ497653*; 11, Montenegro, B. Frajman 17907 (IB), –; 12, Kosovo, N. Berisha 17859 (IB), –; 13, Kosovo, B. Frajman & P. Schönswetter 13799 (IB), ON511314; 14, Kosovo, N. Berisha 17858 (IB), –; 15, Albania, B. Frajman & P. Schönswetter 14007 (IB), ON511315; 16, North Macedonia, B. Frajman 17768 (IB), PQ497652*; Euphorbia tommasiniana Bertol. T1, Italy, B. Frajman 16943 (IB), ON511374; T2, Italy, B. Frajman 16945 (IB), ON511375; Euphorbia valliniana Belli, France, C. Voisin 17095 (IB), ON511377; Euphorbia variabilis Ces., Italy, B. Frajman & P. Schönswetter 14388 (IB), ON511379.




