Super-Resolution Microscopy Using a Bioorthogonal-Based Cholesterol Probe Provides Unprecedented Capabilities for Imaging Nanoscale Lipid Heterogeneity in Living Cells
Abstract
Despite more than 20 years of work since the lipid raft concept was proposed, the existence of these nanostructures remains highly controversial due to the lack of noninvasive methods to investigate their native nanorganization in living unperturbed cells. There is an unmet need for probes for direct imaging of nanoscale membrane dynamics with high spatial and temporal resolution in living cells. In this paper, a bioorthogonal-based cholesterol probe (chol-N3) is developed that, combined with nanoscopy, becomes a new powerful method for direct visualization and characterization of lipid raft at unprecedented resolution in living cells. The chol-N3 probe mimics cholesterol in synthetic and cellular membranes without perturbation. When combined with live-cell super-resolution microscopy, chol-N3 demonstrates the existence of cholesterol-rich nanodomains of <50 nm at the plasma membrane of resting living cells. Using this tool, the lipid membrane structure of such subdiffraction limit domains is identified, and the nanoscale spatiotemporal organization of cholesterol in the plasma membrane of living cells reveals multiple cholesterol diffusion modes at different spatial localizations. Finally, imaging across thick organ samples outlines the potential of this new method to address essential biological questions that were previously beyond reach.
1 Introduction
Biological membranes are poised to laterally self organize, forming defined platforms required for proper cell functioning. The lipid raft hypothesis postulates that specific lipid-lipid and lipid-protein interactions can drive the formation of laterally segregated nanodomains, highly enriched in cholesterol (chol), in which membrane proteins are retained to perform their physiological functions.[1, 2] Those chol-enriched nanostructures are highly packed, in contrast to the rest of surrounding lipids, which form a more fluid phase. Lipid raft have been described to be involved in practically all biological functions and are associated with various human diseases (i.e., cancer, neurodegenerative diseases).[3] This lateral membrane heterogeneity has been extensively validated and visualized at the microscale using model membrane systems and in silico simulations where the lipid composition is easy to adjust.[4] However, in living cells composed of a broad variety of lipids and proteins, lipid raft remains elusive (or illusive).[5] Since seeing is believing, lack of direct visualization of nanoscale lipid heterogeneity in the plasma membrane (PM) of living unperturbed cells increases the skepticism towards the real existence of lipid raft.[6] The astonishing improvement in optical microscopy resolution over the last decades has permitted visualizing single molecules at the nanometer scale. Current super-resolution fluorescence microscopy (SRM) techniques have, by far, overcome the spatial diffraction-limit and allowed for direct observation of cellular processes with high temporal and spatial resolution in living cells.[7, 8] However, despite strong advances in SRM allowing to trap and visualize biological processes at the nanoscale, direct imaging of lipid nanodomains is hampered by the rapid diffusion of lipids and their short lifetime within in vivo cell membranes.[9] The main drawback is that SRM techniques require longer acquisition times and high probe photostability, which often can only be attained by cell fixation.[10] Although cell fixation works relatively well with proteins it is of limited application when using lipids or lipid probes.[11] Recently, environment-sensitive probes compatible with SRM have been developed and tested in living cells. However, when combined with SRM, these Nile-Red-based probes did not prove the existence of lipid heterogeneity in the plasma membrane of living cells but only a higher degree of order in outward and inward protrusion, suggesting a direct association of curvature with order.[12]Authors could not rule out that the presence of even low amounts of the probes does not impact lipid dynamics and domain formation. Similarly, the use of another solvatochromic dye named Di-4-ANEPP combined with STED nanoscopy has enabled to visualize and quantify nano- and micrometric domain dynamics (>100 nm). However, those solvatochromic dyes only inform about membrane order state without giving accurate information about the lipid and protein composition of those domains.[8] Alternatively, secondary ion mass spectrometry has shown to be a valuable technique to visualize the micro- and nanoscale lateral organization of cellular membranes in situ. Nevertheless, this technique is performed under ultra-high vacuum and requires sample fixation.[13] To date, lipid chemical tools used to study lipid membranes have not addressed the features of lipid raft and chol dynamics in living cells at the nanoscale, and the challenge to develop membrane lipid probes suitable for live-cell SRM remains unachieved.
To address the aforementioned need, we chose to develop a bioorthogonal chol probe (chol-N3) that preserves the natural molecule's physicochemical properties and shows outstanding performance in SRM. Our approach enables direct visualization and characterization of nanoscale lipid heterogeneity at the PM of living unperturbed cells. Chol-N3 facilitates direct imaging of membrane lipid dynamics with high spatiotemporal resolution over extended time scales and areas with an unprecedented level of detail. When combined with fluorescence microscopy, chol-N3 permits 3D deep visualization of chol distribution through thick brain tissue samples. We conclude providing a PM model containing yet unknown features of nanoscale chol dynamics in living cell membranes.
2 Results and Discussion
2.1 Synthesis, Design, and Characterization of the Chol Probe
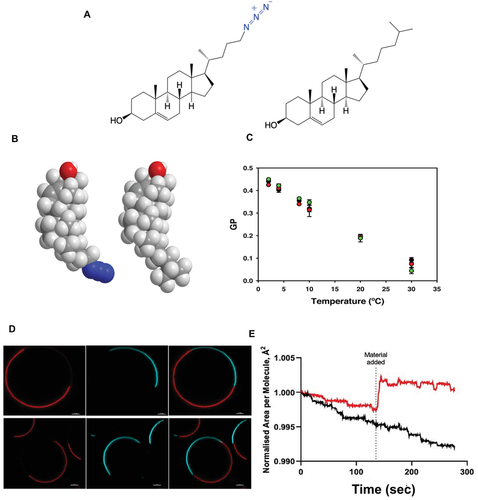
To investigate the existence of chol-rich nanodomains in living cells, we undertook the synthesis of a chol chemical probe, chol-N3, based on bioorthogonal chemistry that can be used to explore many unknown features of lipid raft in living cell membranes. We reasoned that chemical probes for imaging lipid heterogeneity and dynamics should contain a latent chemical modification that minimally perturbs the biophysical and metabolical properties of the chol probe. Bioorthogonal chemistry has shown widespread usage in the past, offering unique advantages over other chemical reactions such as 1) versatility and selectivity, 2) compatibility with all types of biomolecules, and 3) suitability for live-cell imaging. With this idea in mind, we introduced an unobtrusive azide group in position 24 of the chol molecule (Figure 1A). The preparation of chol-N3 is fully described in the Experimental Section. We first started by investigating the natural behavior of the chol probe compared to its endogenous counterpart. In silico experiments show that the absolute stereochemistry and molecular topology in a hydrophobic environment exhibited by chol-N3 are very similar to those observed in unmodified natural chol (Figure 1B). Next, we evaluated the suitability of the chol-N3 probe for imaging purposes in living cells. To this end, living cortical neurons pre-treated with chol-N3 were labeled by copper-free click chemistry using a fluorescently-labeled dibenzocyclooctyne (DBCO) containing a linker between the dye and the reacting group, and were visualized. Confocal imaging of chol-N3 in living neurons demonstrates intense chol-N3 staining, which is particularly robust in the soma, axon, and dendrites (Figure S1 and Movie S1, Supporting Information). Cell labeling with a series of DBCO-linked fluorescence dyes differing in size and polarity showed a similar labeling pattern, ruling out any possible artifact that could be attributed to the fluorescent dye used (Figure S2, Supporting Information). Marginal labeling was observed when cells were grown in the presence of ethanol (vehicle) instead of chol-N3 (Figure S3, Supporting Information). Finally, neither chol-N3 nor the fluorophore-labeled cyclooctyne treatment exert any toxic effect on the cultured cells (Figure S4, Supporting Information). These results validate the use of the new probe for live-cell imaging. Recently, a similar labeling strategy has enabled visualization of mitochondrial phosphatidylcholine containing lipids in living cells by confocal microscopy.[14]
 ), POPC:chol-N3 (90%:10%)(
), POPC:chol-N3 (90%:10%)( ), and POPC:chol-N3:chol-N3-AF 647-DBCO (90%:8%:2%)(
), and POPC:chol-N3:chol-N3-AF 647-DBCO (90%:8%:2%)( ) containing 5 × 10−6 m Laurdan. D) Partitioning characteristics of lipids in phase-separated GUVs composed of DOPC/SM/chol (3:3:1, mol:mol) (top) or DOPC/SM/chol-N3 (3:3:1, mol:mol) (bottom) labeled with 0.2% Rho-DHPE (red, left panel), and 0.5% naphthopyrene (NAP) (blue, middle panel). These probes label the Ld phase and the Lo phase, respectively. Overlay images of the two different channels are shown in the right panel. Scale bar = 5 µm. E) Monolayer experiments at 30 mN m−1. A chol (3.25 × 10−8 mol) monolayer was programmed to maintain a 30 mN m−1 surface pressure prior to adding a 1% (black) of chol-N3-AF647, or a 1% molar ratio of DPPC (red) as a positive control. Area per molecule is normalized to 1% chol-N3-AF647. Increased area per molecule represents an increase of area made to maintain the 30 mN m−1 surface pressure. Lipids addition that minimally changes the area per molecule at 30 mN m−1 indicates that the presence of the dye is not affecting native chol biophysical function. N = 3 independent experiments were completed per condition.
) containing 5 × 10−6 m Laurdan. D) Partitioning characteristics of lipids in phase-separated GUVs composed of DOPC/SM/chol (3:3:1, mol:mol) (top) or DOPC/SM/chol-N3 (3:3:1, mol:mol) (bottom) labeled with 0.2% Rho-DHPE (red, left panel), and 0.5% naphthopyrene (NAP) (blue, middle panel). These probes label the Ld phase and the Lo phase, respectively. Overlay images of the two different channels are shown in the right panel. Scale bar = 5 µm. E) Monolayer experiments at 30 mN m−1. A chol (3.25 × 10−8 mol) monolayer was programmed to maintain a 30 mN m−1 surface pressure prior to adding a 1% (black) of chol-N3-AF647, or a 1% molar ratio of DPPC (red) as a positive control. Area per molecule is normalized to 1% chol-N3-AF647. Increased area per molecule represents an increase of area made to maintain the 30 mN m−1 surface pressure. Lipids addition that minimally changes the area per molecule at 30 mN m−1 indicates that the presence of the dye is not affecting native chol biophysical function. N = 3 independent experiments were completed per condition.To assess whether the chemical modification introduced in this probe interferes with the native behavior of the natural compound, and to verify that decoupling the dye from the molecule by a linker buffers the fluorophore's overall impact on lipid behavior, we carried out a series of control experiments designed to compare the biophysical behavior of chol-N3 (nonlabeled and labeled) to that of chol in model and cell membranes. First, we used the solvatochromic fluorescent probe Laurdan to detect changes in membrane lateral packing properties due to its ability to sense the polarity of the surrounding environment.[15, 16] To this end, we prepared large unilamellar vesicles of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) containing Laurdan in the presence of chol, chol-N3, or chol-N3 labeled with Alexa Fluor 647-DBCO (AF 647-DBCO). As observed in Figure 1C, the generalized polarization value (GP)[17] shows the same membrane condensation or order behavior irrespective of the chol analog used. Next, we investigated the ability of chol-N3 to induce lipid membrane domains in model membranes. For this purpose, giant unilamellar vesicles (GUVs) composed of the ternary mixture POPC/Sphingomyelin (SM)/chol, known to phase separate into liquid-disordered (low chol content, Ld) and liquid-ordered (high chol content, Lo) domains, were used.[18] To examine the partitioning of endogenous chol as well as chol-N3 in GUVs, Rhodamine-PE and naphthopyrene were used as Ld and Lo markers, respectively. As Figure 1D shows, similar phase separation and domain formation were observed with chol and chol-N3. Importantly, when the labeling experiments were carried out in situ using Alexa Fluor 488-DBCO (AF 488-DBCO), the fluorescently-labeled chol-N3 showed strong localization in the Lo phase (Figure S5, Supporting Information), probing that the lipid labeled using a dye with a linker has an identical phase separation to the native form. Finally, we performed Langmuir Trough interfacial experiments to quantify the area per molecule occupied by our dye at meaningful membrane surface pressures (≈30 mN m−1).[16] Addition of dye at the interface showed minimal displacement of the area ocupied by a single dye in comparison with dipalmitoylphosphatidylcholine (DPPC), a saturated lipid forming Lo domains, indicating that the labelled chol occupied a smaller area than a DPPC molecule and could be accomodated without monolayer area disruption (Figure 1E). Altogether, these results point to similar lateral lipid packing distribution, chain order, and membrane organization in model membranes containing either chol or its chemical analog chol-N3, both nonlabeled or labeled with Alexa Fluor dyes containing a linker separating the lipid from the reporter dye. Finally, we investigated whether chol-N3 and chol follow the same intracellular uptake pathway. To this end, we performed an in vivo competition assay in cells where the concentration of chol-N3 was kept constant while increasing the concentration of free chol in the media. As observed in Figure S6 (Supporting Information), increasing levels of extracellular chol block the uptake of chol-N3 gradually, which is virtually abolished at 500 × 10−6 m chol. When the same experiment was repeated with chol-N3 previously labeled with AF 488-DBCO, identical results were obtained to those observed with the unlabeled lipid analogue (Figure S7A, Supporting Information), thus confirming that chol-N3 (labeled or unlabeled) follows the same intracellular uptake pathway as chol in vivo. Strikingly, when the commonly used BODIPY-chol, where the fluorophore is directly linked to the lipid, was used instead of labeled or unlabeled chol-N3, no competition effect was observed (Figure S7B, Supporting Information), suggesting a different cellular uptake mechanism between the BODIPY-labeled lipid and the parental molecule. Overall, the performed experiments show that chol-N3, labeled or nonlabeled, behaves like its endogenous counterpart and is highly suitable for live-cell imaging. Notably, the results prove that dye labeling to the chol backbone by means of a linker does not change the physical/chemical properties of the host lipid. These results are in complete agreement with previous studies showing that the presence of a linker between the target lipid and the dye minimizes the dye's impact on lipid's physicochemical properties.[19]
2.2 Unraveling Nanoscale Lipid Heterogeneity in Resting Living Cells
Having validated the new chol probe, we started imaging chol-rich nanodomains in resting living cells. Hence, we combined our chemical probe with stimulated emission depletion (STED) fluorescence nanoscopy, a broadly used SRM technique to spatially resolve in higher detail (≈50 nm) biological processes at the nanoscale.[20] Living cells treated with chol-N3 were labeled with AF 488-DBCO and visualized by 2D STED nanoscopy. Although fluorescent labeling of chol-N3 before cellular membrane addition also shows great properties as previously demonstrated, it is important to mention that the major advantages of labeling the lipid after cellular uptake are 1) the lipid can be inserted, distributed along cellular membranes, and fluorescently labeled at different time points and 2) since the fluorescent-labeled dye is cell nonpermeable, it allows to label only PM chol and to image it for a short time, removing chol fluorescence coming from intracellular structures, a process that cannot be avoided when an already fluorescently labeled lipid is administrated to cells. As Figure 2A,B shows, resolution of chol spatial distribution along the PM of living cells is highly improved in STED compared to standard confocal microscopy (LSCM) and enables high detail observation of nanoscale lipid heterogeneity in the plasma membrane of living cells. Close-up images (Figure 2C) reveal nanodomains with ∼20–40 nm size as quantified by measuring the “Full Width at Half Maximum” (FWHM) value of the fluorescent intensity line profiles (Figure 2D). Statistical analysis of the chol-enriched nanodomain size distribution by measuring their lateral width[21] revealed two well-defined Gaussian peaks at 48 and 157 nm, and a broad single peak distribution in the domain shape, measured by the lateral sphericity (Figure 2E,F). Overall, these indicate that chol-rich nanodomains are highly heterogeneously distributed across the PM and differ in size and morphology. Similar lipid heterogeneity with sizes well below the diffraction-limit was observed when primary neuronal cells were studied (Figure 3A–C). To ensure that lipid heterogeneity observed in Figure 2 was not an effect of dye aggregation due to the probe and reporter molecule concentrations used, we repeated the same experiment progressively decreasing the concentration of chol-N3 and AF 488-DBCO. As observed in Figure S8 (Supporting Information), even a twenty-fold reduction of both molecule´s concentration allows the visualization of similar highly STED-resolved punctuate chol-rich nanodomains in living cell surfaces. When the same experiment was repeated using two different dyes differing in structure (AF 488 is based on a fluorescein scaffold, whereas AF 555 and AF 647 use cyanines as building blocks) and degree of hydrophobicity (Figure S9A,B,C Supporting Information), similar lipid heterogeneity was observed when cells were visualized by STED nanoscopy (Figure S9D,E, Supporting Information). Finally, to validate that the imaged lipid heterogeneity corresponds to GPI-associated lipid nanodomains, we performed two-color SRM in living cells using the chol probe combined with a classical lipid raft marker, the glycosylphosphatidylinositol-anchored (GPI-anchored) protein CD59, a protein involved in signaling processes.[22] Cells transfected with CD59-SNAP were treated with the chol probe. Protein and lipid were labeled in situ using cell SNAP-Surface AF 647 and AF 488-DBCO, respectively, and visualized by LSCM and STED microscopy (Figure S10A–C, Supporting Information). As observed in Figure S10D (left panel, Supporting Information), the colocalization map shows a strong fluorescent signal correlation between the CD59 and the chol molecule. This high degree of colocalization was further confirmed when the colocalization coefficient was calculated (Figure S10E, left panel, Supporting Information). Considering that the maximum colocalization coefficient obtained for chol imaged in the confocal versus STED mode was 0.814 (Figure S10D,E, right panels, Supporting Information), the results show that more than 75% of GPI-anchor CD59 protein localize into chol-enriched nanodomains in the cell surface of resting living cells. The above results confirm that the lipid heterogeneity observed in Figure 2 is a direct visualization of GPI-associated chol-enriched nanodomains in the plasma membrane of living cells.
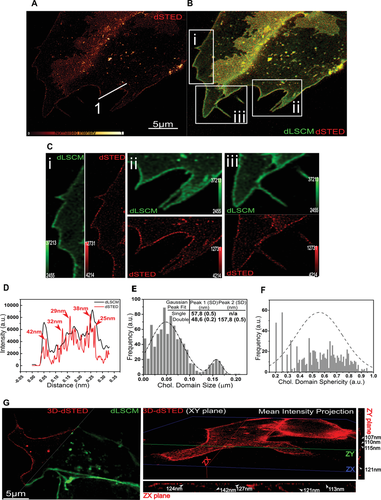
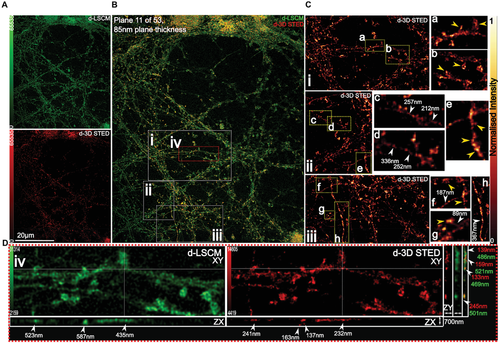
Given the high lateral resolution and probe photostability achieved with 2D STED, we asked whether the plasma membrane's lateral organization could also be super-resolved in the axial plane. This three-dimensional spatial arrangement is critical to fully determine nanodomains morphologies and identify cellular factors (i.e., cytoskeleton) regulating PM lipid heterogeneity. To test this, we combined chol-N3 with 3D STED nanoscopy. 3D STED recording of living cells showed that lipid nanodomains could be super-resolved in the axial plane (Figures 2G and 3D). Figure 2G (right panel) and Figure 3C,D show that lateral lipid organization could be resolved laterally down to 89 nm and axially down to 107 nm, in contrast to deconvolved confocal (d-LSCM) images that showed domain sizes of ≈180 and ≈600 nm, respectively. To the best of our knowledge, this is the first time that chol-rich nanodomains have been directly observed within the membrane of living unperturbed cells with such high resolution in all three planes (XY resolution ≈ 25–35 and ≈110 nm in the Z-axis) and in accordance with their predicted lateral size in living cells.[23]
2.3 Revealing Lipid Heterogeneity and Nanoscale Dynamics in Living Cell Membranes
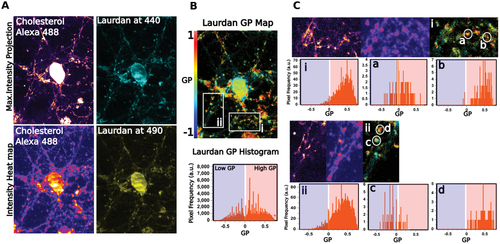
Next, we tried to unravel the membrane lipid structure of such lipid nanodomains in living cells. Lateral organization of the cellular membrane is known to be formed by the clustering of specific lipids into different domains, which are immiscible among them. Highly-condensed domains are differentiated from the surrounding membrane by their lipid structure (Lo vs Ld).[1, 24, 25] To study if the previously observed nanoscale lateral organization in the PM corresponds to Lo domains as in model membranes, we combined the chol probe with Laurdan, a solvatochromic lipophilic probe highly sensitive to changes in the lipid lateral packing properties and the gold-standard of lipid order quantification. This polarity sensitive probe partitions ubiquitously into both phases, shifting its spectral emission properties when it is localized in a more densely packed lipid membrane.[26, 27] Cells were treated in vivo with the chol probe combined with Laurdan. After imaging, the GP map was calculated, and the resolved GP values were displayed as a histogram (Figure 4A,B). The GP map, as well as the distribution of GP values, clearly show nonarbitrary differences in GP values across the cell surface. However, areas with high GP values, indicating a condensed structure, colocalize with the chol-enriched nanodomains labeled with the chol probe (Figure 4C). Notably, Figure 4C also shows that chol-enriched nanodomains also exist in more disordered regions. However, when a large number of chol-rich nanodomains were analyzed, the majority of those nanodomains show a Lo distribution (4.3 ± 0.093 Lo-like domains per micron2 with an average area of 0.003 ± 0.0008 per micron2). Since the emission of Laurdan partially overlaps with the absorption of AF 488, we checked that FRET between the two dyes was not taking place, which could affect GP values. As observed in Figure S11A,B (Supporting Information), neither a significant quenching of AF 488 to Laurdan emission in ethanol or liposomes, nor important variation in GP values in liposomes at two different temperatures, were detected ruling out any significant FRET between the two dyes. Besides, when the same living cell experiment was repeated using Laurdan and AF 647-DBCO, whose emission and absorption spectra are well-separated, identical results were obtained, showing a strong colocalization between the chol labeled probe and the Laurdan ordered channel at 440 nm (Figure S11C–F, Supporting Information). These results demonstrate that as hypothesized in lipid raft, chol-rich nanodomains laterally segregate into condensed lipid structures that differ in the degree of fluidity from the rest of the PM in living cells. It is important to mention that the strategy used here has a major advantage over other approaches that have been used to monitor membrane lipid order.[12, 24] Unlike studies that only used a single probe to analyze plasma membrane lipid compartmentalization, the combination of chol-N3 and a solvatochromic probe used here enables us not only to probe nanoscale lipid heterogeneity in living cells in an unbiased way, but also to ascertain that the Lo structures observed are highly-enriched in chol; the key constituent of lipid raft.
2.4 Nanoscale Spatiotemporal Diffusion Dynamics of chol-N3 in Living Cells
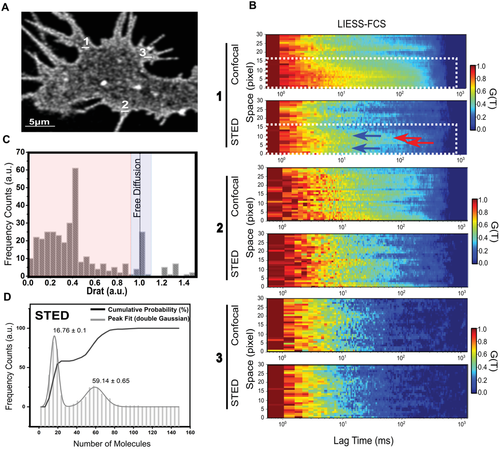
An additional long-standing question in membrane biology is to unravel the nanoscale spatiotemporal organization of lipids within the PM. In other words, is chol diffusion through these ordered nanodomains slower than diffusion along more fluid areas at the PM? We addressed this experimental challenge by combining the chol probe with line interleaved excitation scanning STED Fluorescence Correlation Spectroscopy (LIESS-FCS) in live cells, as recently described.[28] Molecular diffusion dynamics at different spatiotemporal localizations can be measured by LIESS-FCS in one single measurement using fast and multiple scanning across a line (alternating STED and confocal laser illumination) with minimal invasion (Figure 5A). The ratio between the diffusion coefficient by STED and confocal yields the Drat value. As previously reported,[28] a Drat value close to 1 indicates free diffusion, whereas Drat values <1 are characteristic for trapped molecules. Chol diffusion was calculated in living cells at different PM localizations and the corresponding autocorrelation function for each focal spot was used to build up the correlation carpets in either confocal or STED nanoscopy (Figure 5B). Interestingly, two different diffusion modes were observed for chol across the same zone (dotted white-line frame). When the chol transit time is similar within a diffraction-limited (STED) and a larger (confocal) illumination spot, then Drat is lower than 1, thus suggesting a trapped molecular diffusion (Figure 4B, red arrows) with a lipid-specific transient trapping time between 4 and 12 ms and an average diffusion coefficient of D = 0.13 ± 0.04 µm2 s−1. In addition, we also observe a 500 nm region (dotted white-line frame), where the chol resident time was shorter in STED mode (Figure 5B, blue arrows). In this case, the Drat ≈1 value strongly suggests a free diffusion mode for chol with an average diffusion coefficient of D = 1.26 ± 0.05 µm2 s−1. Overall, chol transit time quantification shows a chol spatial and temporal heterogeneity where chol in free and trapped diffusion modes coexist within a nanoscale area. These dual slow chol diffusion modes have been previously observed only using in silico methods, but never in live cells showing visual presence of lipid nanodomains.[29] Reassuringly, our results indicate that the existence of high-ordered and less ordered lipid nanodomains in vivo is highly dependent on the chol content (Figure 5C,D). It is important to mention that when a larger number of chol-enriched areas are analyzed, the most abundant chol diffusion mode is the trapped mobility, as the one depicted in the histogram of Figure 5C. Finally, we determined the average number of chol labeled molecules trapped in ordered lipid nanodomains and freely diffusing through a less-ordered nanodomain within the PM. This is a significant challenge in cell biology, where the concentration and aggregation state of lipids may differ in various localizations and change during biological processes. To this end, we quantified the molecular brightness[30] allowing us to calculate the apparent average number of molecules in each pixel from our series of LIESS-FCS-STED measurements. From our STED measurements, we found an average number of 59.14 ± 0.65 molecules of confined chol-labeled probe and 16.76 ± 0.1 molecules freely diffusing through the investigated area (Figure 5D). Since potential background signal (e.g., noise, internalized fluorescence) can affect molecular brightness measurements, we tested whether chol-N3 signal in the PM was stable over time. As observed in Figure S12A, (Supporting Information), chol-N3 labeled with AF488-DBCO fluorescence signal in the PM is very stable with no photobleaching or loss of fluorescence over long-time measurements. However, fluorescence intensity fluctuations were observed in intracellular vesicles filled with chol-N3 (Figure S12A, Supporting Information). To confirm these results, we performed raster imaging correlation spectroscopy (RICS) experiments of the chol-N3 probe in the PM of living cells. RICS allows tracking biomolecule diffusion in living cell membranes in space and time simultaneously at large regions of interest.[8, 31] Hence, it enables us to move across wide length and temporal scales. As Figure S12B, Supporting Information, shows chol-N3 labeled with AF488-DBCO fluorescence intensity is very stable over time and fast and slow chol-N3 diffusions through the PM can be detected in agreement with our LIESS-STED-FCS results. These findings shed new light into the complex heterogeneous nature of cell membranes in real-time, showing that while the majority of chol is trapped into ordered nanodomains, chol is also found to form less-ordered and diffusible domains through the PM of living cells.
2.5 3D Volumetric Mapping of Chol Distribution in Intact Tissue
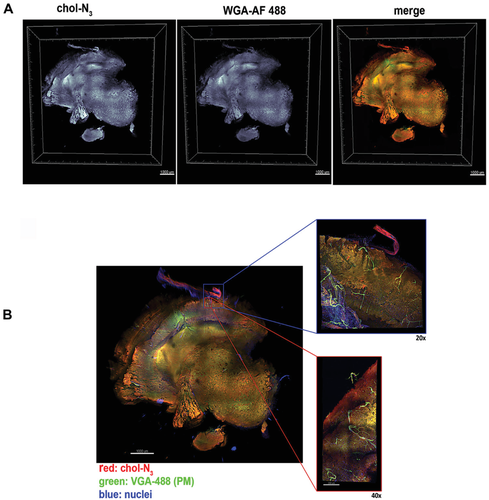
Visualizing single lipid species at high-resolution in tissues has been difficult, especially in the brain, where large concentrations of lipids and chol can be found. We tried to map chol distribution above the cellular level, by studying if high-resolution images of tissues or even complete organs could be obtained. Our starting hypothesis was that given the chol probe's unique properties in living cells, this could help to visualize the 3D chol organization in thick brain samples by fluorescence microscopy without the need to slice the sample into thin serial sections. For this purpose, thick rat brain slices, varying in thickness from 500 µm to 1 mm, were labeled ex vivo with the chol probe. Figure 6A shows a 3D volumetric reconstruction of thick brain slices where the cells were densely and homogenously labeled throughout the sample. These results are in good agreement with the expected general chol distribution through brain cells (i.e., oligodendrocytes and astrocytes).[32] To further corroborate the localization of chol-enriched structures in the plasma membrane, we treated the brain samples with a fluorescently labeled lectin, a well-known plasma membrane marker. We found a strong colocalization between the chol probe and the plasma membrane marker (Figure 6A; Movie S2, Supporting Information). On the other hand, in the absence of chol-N3 treatment, no fluorescence labeling through the tissue was observed (Figure S13, Supporting Information). Finally, we tried to determine to what extent the high-resolution visualization of chol in deep-tissue structures was possible. To this end, representative sections of rat brain cortex were imaged at high magnification by confocal microscopy. With different objectives, we were able to obtain 3D volumetric reconstructions of magnified areas where the distribution of chol in brain cells is clearly shown (Figure 6B).
Overall, through the design and development of a bioorthogonal-based chol probe that retains similar physicochemical properties compared to the parental molecule when fluorescently labeled and performs superbly in SRM, we have provided compelling evidence about the existence of lipid raft in the cell surface of living unperturbed cells, a long-lasting mystery in science. Using the chol probe we could answer many remaining biological questions about lipid nanoscale heterogeneity in cells. We have recapitulated these new biological data in a PM model (Figure 7). When the probe was combined with SRM in living cells, we could directly observe and characterize PM nanoscale heterogeneity with domain sizes far below the diffraction limit in all three planes (XY ≈ 25–35 nm with 2D STED and ≈ 110 nm in the Z-axis, 3D-STED) (Figures 2 and 3). Compared to other methods that supported the existence of lipid raft by indirect evidence, either tracking fluorescently labeled proteins that, supposedly, populate such lipid structures[33] or by chol depletion using pharmacological drugs and chemicals as methyl-β-cyclodextrins, which seems to affect cellular architecture,[34] here we directly observed their native nanoscale organization in the PM of living cells in a noninvasive manner. Performing dual-color live-cell SRM, we observed a strong enrichment of a classical lipid raft marker, the GPI-anchor protein CD59, with chol in the cell surface of living cells (Figure S10, Supporting Information). A previous study using protein micropatterning combined with single-molecule tracking of GFP-tagged GPI anchor proteins showed that GPI-anchor proteins do not reside in Lo domains in the cell surface of cells.[35] However, recent works using a combination of fluorescent lipid nanodomain markers and single-molecule tracking obtained similar results to our work where GPI-anchor proteins colocalize with a classical lipid raft marker, the ganglioside GM1, for different periods in the PM of living cells.[36] These contradictory results can be explained by the fact that in micropatterning experiments, GFP-GPI anchor proteins expressed in cells are immobilized to streptavidin enriched coverslips using a biotinylated GFP antibody, and this process may change the protein's physicochemical properties and thus affect how the protein interacts with other PM components and the cytoskeleton. When we combined the chol probe with an environment-sensitive probe, we ascertained that the lipid structure of the observed nanodomains was mainly a Lo structure (Figure 4; Figure S11, Supporting Information). Those results are in accordance with previous studies that determined using environment-sensitive probes and SRM that the PM is mainly composed of 76% ordered and 24% disordered lipid domains.[27] However, in our living cell experiments, a highly complex picture of cellular membrane organization emerged, where distinct chol-enriched nanodomains with different degrees of order coexist. This has been previously observed in model membranes[37] but not in live cells. The LIESS-FCS-STED experiments with the probe shed new light on the organization of chol-rich nanodomains by revealing different degrees of chol lateral mobility in real-time within the membrane depending on its partition between highly-ordered (where chol is trapped with a retention time >4 ms) and less-ordered domains (where chol diffuses freely with retention times <4 ms). However, as observed with the lipid structure experiments, when a large number of nanodomains were analyzed, the majority of nanodomains showed a Lo structure and trapped state (Figure 4). Interestingly, similar retention times (>4 ms) to the ones found in this work were observed using scanning STED-FCS for lipids showing a trapped mode.[38] Quantifying the molecular brightness from the LIESS-FCS-STED recordings enables us to determine the number of chol labeled molecules present in an ordered (≈60 molecules) and less ordered (≈17 molecules) nanodomain.

A striking example that could not be answered without the chol-N3 probe is provided by the generation of high-resolution 3D volumetric images showing the distribution of chol through thick brain sections by confocal microscopy in an unprecedented manner and highlights its versatility for tissue imaging. This methodology could be further extended to investigate the effects of chol on tissue or organ development in vivo by fluorescent microscopy. The approach described in this work has some limitations that have to be considered. For instance, the molecule has to be administrated exogenously to cells, and labeling using a reporter dye containing a linker could change some properties of the labeled lipid. While our strategy using a linker between the lipid and the reporter molecule buffers the impact of the dye in the physicochemical properties of chol (Figure 1; Figure S5, Supporting Information), enabling us to track chol at the nanoscale (Figures 2, 3, and 5), we can not rule out a direct effect of the labeling at the atomistic level. In this sense, molecular dynamics simulation studies could clarify whether the reporter molecule's presence alters how the labeled lipid interacts with surrounding biomolecules at the atomistic level. Finally, due to the versatility of click chemistry, the chol-N3 present within cells and tissues can also be modified by any molecule bearing an alkyne or cyclooctyne group that could serve as a functional tool for future in vivo studies using a wide range of techniques, such as cryo-correlative light and electron microscopy and magnetic resonance imaging.
3 Conclusions
In this work, we developed chol-N3, a bioorthogonal-based chol probe that, when combined with SRM, provides direct visualization of nanoscale lipid heterogeneity in the cell surface of resting living cells. The chol-N3 offers a new dimension to study the impact of lipid raft in fundamental biological processes in living cells with high spatial and temporal resolution over extended time scales and areas. For instance, chol-N3 can be combined with a wide range of dyes compatible with live-cell SRM imaging or with fluorescently labeled biomolecules (i.e., proteins). This strategy proficiently facilitates the study of the implication of nanoscale lipid heterogeneity in a large number of cellular processes such as receptor signaling, trafficking, and unraveling the role of actin as a direct orchestrator of plasma membrane compartmentalization since it offers the unique ability to track biomolecules by dual-color live-cell SRM directly. Finally, this bioorthogonal probe opens up new avenues for the in-depth investigation of chol tissue organization and its impact on health and disease.
4 Experimental Section
The following lipids were purchased from Avanti lipids (Alabaster, AL): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOTAP), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), Bodipy-chol (TopFluor-chol) and cholesterol (Chol). Lissamine Rhodamine B 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine (Rho-PE) and Cholera Toxin B conjugated to Alexa Fluor 647 were purchased from ThermoFisher Scientific (Waltham, Massachusetts, Ma). Naphtho[2,3a-]pyrene (NAP) was purchased from Sigma-Aldrich (St. Louis, MO). Alexa Fluor 488-DBCO (AF 488-DBCO), Alexa-Fluor 555-DBCO (AF 555-DBCO), Alexa-Fluor-647-DBCO (AF 647-DBCO) and sulfo-Cyanine3-DBCO (sulfo-cy3-DBCO) were purchased from click chemistry tools (Scottsdale, AZ). pME-puro-SNAP-FLAG-CD59 was a gift from Reika Watanabe (Addgene plasmid # 50374; http://n2t.net/addgene:50374; RRID:Addgene_50374).
Synthesis of Compounds—General Procedures
All reagents were obtained from Sigma-Aldrich, Fisher Scientific, or TCI and used without further purification. Anhydrous N,N-dimethylformamide, Tetrahydrofuran, dichloromethane were purchased in sealed bottles and stored over molecular sieves. Deuterated solvents were purchased from Fisher Scientific. Solvents were removed on an IKA Rotavapor RV10 equipped with an ILMVAC LVS210TE vacuum pump. Thin layer chromatography was performed with silica 60 Å gel TLC plates from Merck (60 F254) and analyzed by UV illumination or 20% sulfuric acid staining solution. High-pressure NMR spectra were acquired on Bruker AV-500. 1H NMR spectra were referenced to residual CHCl3 (7.26 ppm). 13C NMR spectra were referenced to CDCl3 (77.16 ppm). NMR spectra were processed using MestReNova (Mestrelab Research S.L.). High-resolution ESI mass spectra of small molecules were obtained at the University of the Basque Country (EHU/UPV) Mass Spectrometry Facility on a Thermo LTQ Orbitrap mass spectrometer.
Synthesis of Compounds—Experimental Procedures
Synthesis of 4-(3-((tert-butyldimethylsilyl)oxy)-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanoic acid 1): Compound 1 was synthesized as previously described in [39] with slight modifications. Briefly, in a three-neck bottom flask containing a solution of 3β-hydroxy-5-cholenic acid (0.305 g, 0.814 mmol) in DMF (7 mL) at 0 °C was subsequently added Imidazole (0.445 g, 6.54 mmol), DMAP (0.200 g, 1.64 mmol), and TBDMSCl (0.370 g, 2.45 mmol). The solution was stirred overnight at room temperature. After stirring overnight, the reaction was quenched with 1 m HCl (5 mL) and extracted with EtOAc. The organic layer was washed with brine dried over MgSO4, and the solvent was removed under reduced pressure. Next, the crude product was dissolved in 14 mL MeOH/THF (1/1, v/v) and a 10% (w/w) K2CO3 (4 mL) solution was added in water. After stirring for 1 h at room temperature, the reaction mixture was concentrated under reduced pressure. The product was resuspended in 25 mL brine and acidified to pH 3 with 1 m HCl. Finally, the compound was extracted with EtOAc (3 × 50 mL), dried over MgSO4 and concentrated in vacuo. The crude product was purified using silica gel chromatography (hexanes: diethyl ether: acetic acid, 70:30:1 v:v:v) to afford a white solid. Yield: 0.345 g (87%).
1H NMR (CDCl3, 500 MHz) δ = 5.32 (m, 1H), 3.49 (tt, J = 10.9, 10.9, 4.7, 4.7, 1H), 2.40 (m, 1H), 2.31– 2.23 (m, 2H), 2.17 (m, 1H), 1.98 (m, 2H), 1.91–1.67 (m, 4H), 1.64–1.24 (m, 10H), 1.20–1.03 (m, 4H), 1.00 (s, 3H), 0.94 (d, J = 6.5, 3H), 0.89 (s, 9H), 0.68 (s, 3H), 0.06 (s, 6H). 13C NMR (CDCl3,126 MHz): −4.59 (x2), 11.86, 18.26, 18.29, 19.42, 21.05, 24.25, 25.94 (x3), 28.09, 30.78, 30.90, 31.91 (x2), 32.08, 35.31, 36.58, 37.39, 39.77, 42.39, 42.81, 50.17, 55.75, 56.77, 72.65, 121.12, 141.57, 179.83 ppm. HRMS (ESI) calcd for C30H52NaO3S: 511.3583 [M+Na]+; found: 511.3576.
Synthesis of 4-(3-((tert-butyldimethylsilyl)oxy)-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentan-1-ol (2): Compound 2 was synthesized as described in[39] with minor modifications. In a 100 mL bottom flask containing a slurry solution of LiAlH4 (0.120 g, 3.16 mmol) in THF (5 mL) at 0 °C was added dropwise a solution of (1) (0.340 g, 0.6962 mmol) in THF (20 mL). The reaction mixture was allowed to warm to room temperature and stirred for 1.5 h. Next, at 0 °C the reaction mixture was quenched with subsequently dropwise addition of H2O (110 μL), 15% NaOH (110 μL), and H2O 322 μL. After 1 h, the granular salt was filtered off, the organic layer washed with brine, dry over MgSO4 and concentrated in vacuo. The crude product was purified by column chromatography (hexanes: EtOAc (85/15 v:v) to afford a withe yellowish solid. Yield: 0.265 g (80%).
1H NMR (CDCl3, 500 MHz) δ = 5.32 (dt, J = 4.5, 2.0, 2.0, 1H), 3.49 (m, 1H), 2.40 (m, 1H), 2.27 (m, 2H), 2.17 (m, 1H), 2.02–1.94 (m, 2H), 1.91–1.77 (m, 2H), 1.75–1.64 (m, 1H), 1.63–1.33 (m, 7H), 1.33–1.24 (m, 8H), 1.00 (s, 3H), 0.94 (d, J = 6.6, 4H), 0.89 (s, 9H), 0.68 (d, J = 1.7, 4H), 0.09 (s, 1H), 0.08 (d, J = 0.9, 1H), 0.06 (s, 6H). 13C RMN NMR (CDCl3, 126 MHz,) δ = 141.72, 121.27, 72.80, 56.91 × 2, 55.90, 50.32, 42.96, 42.54, 39.92, 37.54, 36.73, 35.46, 32.22, 32.05 (x2), 29.84, 28.24, 26.09 (x3), 24.40, 21.20, 19.57, 18.88, 18.44, 18.41, 12.01, −4.44 (x2).
HRMS (ESI) calcd for C30H54O2SiNa: 497.3791 [M+Na]+; found: 497.3783.
Synthesis of 4-(3-((tert-butyldimethylsilyl)oxy)-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentyl methanesulfonate (3).To a solution of (2) (0.270 g, 0.49 mmol) in dry CH2Cl2 (25 mL), neat ET3N (0.075 g, 0.74 mmol) was added dropwise at 0 °C. The resulting mixture was vigorously stirred for 15 min, followed by dropwise addition of neat MsCl (0, 085 g, 0.74 mmol). After stirring for 3 h at 0 °C, the reaction mixture was diluted by addition of CH2Cl2 (10 mL). The organic layer was successively washed with 0.5 n HCl (20 mL), aqueous sat. NaHCO3 (20 mL), and brine (20 mL), dried over MgSO4, filtered and concentrated under reduced pressure. The crude product was purified by flash chromatography (hexane/ethyl acetate 2/1) to afford compound 3 as a white powder. Yield: 0.260 g (96%).
1H NMR (CDCl3, 500 MHz) δ = 5.32 (m, 1H), 4.23–4.18 (m, 2H), 3.48 (m, 1H), 3.00 (s, 3H), 2.27 (m, 1H), 2.17 (m, 1H), 2.03–1.94 (m, 2H), 1.88–1.76 (m, 3H), 1.74–1.38 (m, 8H), 1.26 (s, 6H), 1.16–1.06 (m, 3H), 1.00 (s, 3H), 0.95 (d, J = 6.5, 3H), 0.89 (s, 9H), 0.68 (s, 3H), 0.06 (s, 6H). 13C NMR (CDCl3, 126 MHz) δ = 141.57, 121.10, 72.62, 70.66, 56.77, 55.99, 50.16, 42.81, 42.36, 42.34, 39.77, 37.40, 37.38, 36.57, 35.29, 32.08, 31.89, 31.52, 29.69, 28.20, 25.83, 24.23, 22.69, 19.42, 18.55, 11.85, −4.59. HRMS (ESI) calcd for C31H56NaO4SSi: 575.3566 [M+Na]+; found: 575.3558.
Synthesis of 17-(5-azidopentan-2-yl)-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-ol (4). To a three-necked round, bottom flask fitted with a reflux condenser was added a solution of (3) (0.200 g, 0.36 mmol) in anhydrous DMF (5 mL). To the resulting solution, NaN3 (0.094 g, 1.44 mmol) was added portion wise under argon atmosphere, and the resulting mixture was stirred overnight at 80 °C. After cooling down to room temperature, the resulting mixture was quenched by addition of water (20 mL), and the aqueous phase extracted with Et2O (3 × 25 mL). The combined organic phases were dried over MgSO4, filtered and evaporated in vacuo to give a crude mixture, which was purified by gel chromatography (hexanes: diethyl ether: acetic acid, 70:30:1 v:v:v) to afford a white powder. The product was dissolved in 20 mL THF and 4 mL 4 n HCl was added dropwise to the solution. After 2 h, the deprotection was stopped by addition of an excess of Na2CO3. The reaction mixture was diluted with CH2Cl2 (30 mL), washed with 0.1 m HCl (20 mL), brine (20 mL) and dried over MgSO4, filtered and concentrated under reduced pressure. The crude product was purified by flash chromatography (hexane/ethyl acetate 3/1) to afford compound 4 as a white powder. Yield: 0.120 g (92%).
1H NMR (CDCl3, 500 MHz) δ = 5.35 (m, 1H), 3.53 (tt, J = 11.1, 11.1, 4.6, 4.6, 1H), 3.23 (m, 2H), 2.33–2.20 (m, 2H), 1.98 (m, 2H), 1.84 (m, 3H), 1.71–1.40 (m, 8H), 1.31–0.90 (m, 16H), 0.69 (s, 3H). 13C NMR (CDCl3, 126 MHz) δ = 140.74, 121.66, 71.77, 56.71, 55.84, 51.95, 50.07, 42.33, 42.25, 39.73, 37.22, 36.47, 35.44, 32.90, 31.86, 31.61, 28.19, 25.48, 24.23, 21.04, 19.37, 18.62, 18.38, 11.84 ppm. HRMS (ESI) calcd for C24H39N3NaO: 408.2991 [M+ Na]+; found: 408.2995.
Minimize Molecular Modeling Simulations
Lowest energy structures in vacuum of molecules were computed using CS Chem3D Ultra software employing the MMF94-force field and the steepest-descent algorithm. Minimum RMS gradient was set to 0.1; minimum and maximum move to 0.000001 and 1.0, respectively. Maximum number of interactions moves to 5000.
In Vitro Laurdan Experiments
Large unilamellar vesicles (LUV) composed of POPC:chol (90%:10%), POPC:chol-N3 (90%:10%) or POPC/chol-N3/chol-N3-AF 647-DBCO (90%: 8%: 2%) were prepared following the extrusion method described by.[40] Laurdan was added in the organic phase before solvent evaporation. Laurdan labeled liposomes at a concentration of 30 × 10−6 m were kept under continuous stirring, and the determination of GP profiles at different temperatures was performed. All fluorescence measurements were made using an SLM Aminco series 2 (Spectronic Instruments, Rochester, NY) spectrofluorimeter as previously described.[15] To quantify changes in the Laurdan emission spectrum, generalized polarization (GP) values were calculated as follow: GP = (IB-IR)/(IB+IR), where IB (at 440 nm) and IR (at 490 nm) correspond to the intensities at the blue and red edges of the emission maxima, respectively.[41]
Electroformation and Visualization of Giant Unilamellar Vesicles (GUVs)
Giant unilamellar vesicles (GUVs) were prepared using a homemade chamber that allows direct visualization under the microscope.[42] Briefly, DOPC/SM/chol (3:3:1, mol/mol) or DOPC/SM/chol-N3 (3:3:1, mol/mol) were prepared to a final concentration of 0.2 × 10−3 m in chloroform/methanol (2:1, v/v), plus 0.2% Rho-DHPE (marker for Ld phases) and 0.5% of NAP (marker for L0 phases). For in situ chol-N3 labeling, DOPC/SM/chol-N3 (3:1:1, mol/mol) GUVs containing 0.2% Rho-PE and 1% GM1 (ganglioside) were prepared to a final concentration of 0.2 × 10−3 m. Next, 2 µl of the lipid mixture stock was spread on the platinum wires of the electroformation chamber and organic solvent traces removed by placing the chamber under high vacuum for 45 min. Chamber was then equilibrated at 70 °C for 15 min followed by addition of 500 µl of 70 °C preheated Millie Q water. An AC electrical field, 10 Hz, 2.5 Vpp, was then applied for 2 h to the electrodes using a wave generator (TG330 function generator; Thurlby Thandar Instruments, Huntingdon, UK). After switching off the generator, vesicles were left to equilibrate for 30 min at room temperature. GUVs attached to the platinum electrodes were visualized using a Nikon D-eclipse C1 confocal system (Nikon Inc., Melville, NY, USA). For in situ analog labeling, 1% of chol-N3 was labeled in situ with AF 488-DBCO for 30 min. After extensive washing, cholera toxin conjugated AF 647 (3 µg mL−1) was added to the mixture, which binds GM1 and labels the Lo phase.
Langmuir Experiments
The NIMA Langmuir-Blodgett Trough (Coventry, England) was custom designed with a continuously enclosed Teflon-vitrified coated ribbon replacing classical barriers as used elsewhere.[16] Surface pressure was quantified with a soaked Wilhelmy cellulose plate. The trough was filled with 0.9% saline (pH 5.8) and left to reach 25 °C prior to deposition of chol variants. Pressure control experiments were carried out by setting the Langmuir Trough to maintain 30 mN m−1 and adding 3.25 × 10−8 mol chol diluted to 13 × 10−3 m in a mixture of chloroform/methanol. 1% or 2% molar ratios of chol-N3-AF647, or a 1% molar ratio of DPPC as a positive control. To produce the isotherms seen in Figure S6 (Supporting Information), deposition of 0.0126 mg chol-N3 or chol-N3 fluorescently labelled with AF647-DBCO diluted in chloroform-methanol (at 13 × 10−3 and 1.2 × 10−3 m, respectively). Compressions were completed at a rate of 50 cm2 min−1 to produce surface pressure-area isotherms.
Competition Experiments
A competition assay was designed to test chol-N3 and fluorescently pre-labeled chol-N3 uptake by cells. HeLa cells were seeded onto coverslips in DMEM supplemented with 10% FBS, and coincubated with a fixed concentration of chol-N3 (5 × 10−6 m) or with chol-N3 (5 × 10−6 m) labeled with 10 × 10−6 m AF 488-DBCO and an increasing concentration of natural chol (ranging from 0 to 1 × 10−3 m) added externally to the cells. After 1 h coincubation of cells with the mixture, cells were stained in vivo with 5 × 10−6 m AF 488-DBCO for 60 min, counterstained with DAPI, fixed, and imaged using a ZEISS apotome. Alternaltively, HeLa cells were treated with 5 × 10−6 m Bodipy-chol in DMEM supplemented with 10% FBS and coincubated with increasing concentration of natural chol (0–1 × 10−3 m) for 1 h. Cells were fixed, and counterstained with DAPI.
Cytotoxicity Experiments
Cytotoxicity of chol-N3 was evaluated by measuring LDH release in treated HeLa and primary neuron cells with the CytoTOX 96 Non-Radioactive Cytotoxicity Assay (Promega) according to the manufacturer's instructions. Cells were treated with either 15 × 10−6 m chol-N3 or 0.2% ethanol in culture medium, or left untreated for 18 h at 37 °C with 5% CO2. Positive control cell samples were lysed with 10% Lysis Solution for 45’ at 37 °C with 5% CO2. Fresh cell culture medium was used as negative control. Cytotoxicity was calculated as follow: (average sample absorbance at 490 nm – average absorbance of the negative control)/(average absorbance of the positive control 490 nm – average absorbance of the negative control) × 100.
Fusogenic Liposome Preparation
Fusogenic liposomes for effective chol-N3 delivery onto brain tissue were prepared as previously described in.[43] Briefly, liposomal lipid components were mixed in chloroform in a molar ratio DOTAP/DOPE/chol-N3 or DOTAP/DOPE/chol (1/1/0.1). The lipid was deposited as a film on the wall of a glass test tube by solvent evaporation under nitrogen. Solvent traces were removed by placing the sample 1 h in a vacuum chamber. Next, the lipid film was suspended in artificial cerebral spinal fluid buffer (ACSF; in × 10−3 m, NaCl 125, NaHCO3 25, KCl 2.5, NaH2PO4 1.25, glucose 25, CaCl2 2, MgCl2) by vortexing at room temperature to form multilamellar vesicles. Finally, homogenization of the sample in an ultrasonic bath for 15 min produces mainly unilamellar vesicles.
Cell Culture and Maintenance
Human cervix adenocarcinoma HeLa cells and human bone marrow neuroblastoma SH-SY5Y cells were obtained from ATCC. HeLa cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS, 2 × 10−3 m Glutamax, 100 units mL−1 penicillin, and 100 µg mL−1 streptomycin (Life Technologies) at 37 °C with 5% CO2. SH-SY5Y cells were maintained in DMEM/F12 (F12 and Dulbecco´s modified Eagle's medium) supplemented with 10% FBS and 1% penicillin and streptomycin. (Life Technologies) at 37 °C with 5% CO2.
Ethics Statement
Animal husbandry and all experimental procedures involving rats were approved by the animal ethics committee of the University of the Basque Country and by the local authorities (authorization number: CEEA M20-2018-97 and 99) and were conducted in accordance with the Directives of the European Union on animal ethics and welfare.
Preparation and Maintenance of Cultured Neurons
Primary cultures of embryonic rat day 18 (E18) (strain Sprague-Dawley) cortical neurons were prepared by using standard procedures; animal dissection was set as DIV0 (Day In Vitro zero). Briefly, cortical cells were dissociated with 0.25% trypsin and 0.1 mg mL−1 DNAse for 15 min at 37 °C, then resuspended by gentle trituration with a Pasteur pipette in MEM (Minimum Essential Medium) with 10% horse serum and 0.6% glucose. 2.5 × 105 neurons were plated onto poly-L-lysine-coated glass-bottom Petri dishes (Ibidi) or coverslips in MEM-HS-glucose. After 3 h, the medium was replaced with Neurobasal A medium containing B27 supplement and 2 × 10−3 m Glutamax (Invitrogen) and neurons were incubated at 37 °C in a humidified incubator with 5% CO2 during 6 days. At DIV7 and DIV14, 1/5 volume of the medium was replaced with fresh Neurobasal medium with 2% B27. At DIV7 2.5 × 10−6 m cytosine arabinoside was added to the medium.
Live-Cell Imaging
Cortical neurons (DIV14) were seeded onto poly-L-lysine-coated glass-bottom Petri dishes (Ibidi 81158) in media Neurobasal A containing B27 serum. Neurons were labeled with 15 × 10−6 m chol-N3 for 16 h at 37 °C. Neurons were then washed with 3 × 1000 μL PBS, then incubated with 10 × 10−6 m AF 555-DBCO in media for 30 min at 37 °C. Finally, cells were extensively washed to remove nonreacted probe and immediately proceed for imaging by confocal microscopy using either a Zeiss LSM880 with Airyscan and a Plan-Aprochromat 63x/1.40 oil objective, a Leica TCS SP5 with a Plan-Apochromat 63x/1.30 glycerol objective, or a LEICA SP8 3X STED SMD confocal microscope (Leica Microsystems, Manheim, Germany) with an HCX PL APO 63x/1.2NA CORR CS2 water immersion objective. Images were further processed using Imaris imaging software (Bitplane) or Huygens, SVI (Netherlands).
STED Nanoscopy
SH-SY5Y cells were seeded onto imaging dishes (Ibidi 81156) in media DMEM/F12 and treated with 25 or 15 × 10−6 m chol-N3 for 16 h at 37 °C. Next day, cells were washed and subjected to in vivo labeling with different concentrations of AF 488-DBCO, AF 555-DBCO, or AF 647-DBCO for 30 min at 37 °C followed by extensive washing (3 × 1000 μL PBS) to remove unreacted probe and proceed for imaging. For dual-color SRM experiments, SH-SY5Y cells were seeded onto imaging dishes (Ibidi 81156) in media DMEM/F12 and CD59-SNAP tag vector (1µg) were transfected using FuGENE HD following manufacters instructions and treated with 15 × 10−6 m chol-N3 for 16 h. Finally, SNAP-tag CD59 protein and chol-N3 were labelled with cell surface SNAP-tag AF 647 (5 × 10−6 m) and AF 488-DBCO (10 × 10−6 m), respectively for 30 min at 37 °C. Cells were washed extensively (3 × 1000 μL PBS) and visualized immediately by LSCM and STED nanoscopy. Cortical neurons (DIV14) were seeded onto poly-L-lysine-coated imaging dishes (Ibidi 81158) in media (Neurobasal A containing B27 serum). Neurons were labeled with 15 × 10−6 m chol-N3 for 16 h at 37 °C. Neurons were then washed with 3 × 1000 μL PBS, then incubated with 10 × 10−6 m AF 488-DBCO in media for 30 min at 37 °C. Finally, cells were extensively washed to remove nonreacted probe and immediately proceed for microscopy. STED and LSCM images were taken in a commercial LEICA SP8 3X STED SMD confocal microscope (Leica Microsystems, Manheim, Germany) equipped with the STED white 100x NA1, 40 oil objective. The microscope was equipped with 3 depletion lines and for this work; the 592 nm depletion line was used. The excitation laser beam consisted of a pulsed (80 MHz) super-continuum white light laser (WLL). For a cleaner emission, the excitation lines were used a notch filter (NF488) in the optical pathway. The pinhole was set at one Airy unit. The gated HyD detectors were set with the gated option on and the temporal gated selected was from 2 to 6.5 ns when depleting at 592 nm. Imaging and processing have been done employing either commercial (Leica LAXs, Huygens, SVI) or open source software (ImageJ); analysis and graphing, employing EXCEL (Microsoft) or Origin Pro (OriginLab).
In Vivo Laurdan Experiments
Cortical neurons (DIV14) or SH-SY5Y cells were seeded onto poly-L-lysine-coated glass-bottom Petri dishes (Ibidi 81158) in media Neurobasal A containing B27 serum or DMEM/F12 media, respectively. Cells were labeled with 15 × 10−6 m chol-N3 for 16 h at 37 °C. Cells were then washed with 3 × 1000 μL PBS, then incubated with 10 × 10−6 m AF 488-DBCO or 10 × 10−6 m AF 647-DBCO, and 2.5 × 10−6 m Laurdan in media for 30 min at 37 °C. Next cells were extensively washed with PBS (3 × 1000 μL PBS) and imaged on a LEICA SP8 3X STED SMD confocal microscope (Leica Microsystems, Manheim, Germany), with a HCX PL APO 63x/1.2NA CORR CS2 water immersion objective and using solid state laser excitation at 405 nm and measuring the intensity between 420 and 460 nm and 470–510 nm for later Laurdan GP quantification. Over 10 different regions of interest at different areas of the acquired images were analysed using simFCS (G-SOFT Inc., Champaign, IL). GP values were represented frequency histograms and were nonlinearly fitted to two Gaussian distributions using OriginPro (OriginLab, Northampton, MA). Only those fits were considered acceptable for which the chi-square test gave values of p > 0.95. Experiments were performed independently at least three times.
LIESS-FCS Experiments
SH-SY5Y cells were seeded onto imaging dishes (Ibidi 81156) in media DMEM/F12 and treated with 15 × 10−6 m chol-N3 for 16 h at 37 °C. Cells were labelled with 10 × 10−6 m AF 488-DBCO for 30 min at 37 °C and extensively washed with PBS. Line interleaved excitation and stimulated emission depletion FCS (LIESS-FCS) scanning was recorded on a LEICA SP8 3X STED SMD confocal microscope (Leica Microsystems, Manheim, Germany), with a HCX PL APO 63x/1.2NA CORR CS2 water immersion objective using the line step function and alternating the excitation between the confocal and the STED modes between every scanned line, in a similar fashion as described before,[28] but in a line rather than circular scanning. Finally, the obtained intensity data for confocal and STED modes were extracted into two different channels. For this work, STED and confocal observation spot size 100 and 240 nm, respectively, were used. The values of Drat = Dap(STED)/Dapp (confocal) over the x axis were obtained as previously described.[28] Analysis of the data was done employing custom-built software FOCUScan as described before[44] and freely available (see the Supporting Information in the paper). The transit time ratios (Drat) between the diffusion coefficient values obtained from the analysis of the scanning FCS in STED and in confocal mode were graphed as a function of the number of events (occurrence) employing OriginPro (OriginLab, USA).
RICS Experiments
SH-SY5Y cells were seeded onto imaging dishes (Ibidi 81156) in media DMEM/F12 and treated with 15 × 10−6 m chol-N3 for 16 h at 37 °C. Cells were labelled with 10 × 10−6 m AF 488-DBCO for 30 min at 37 °C and extensively washed with PBS. RICS acquisitions were recorded on a commercial LEICA SP8 3X STED SMD confocal microscope (Leica Microsystems, Manheim, Germany), with an HCX PL APO 63x/1.2NA CORR CS2 water immersion objective and using a WLL as pulsed laser source. Relative power, as it appears in the LAXs software was always below 2%. The images shown are representative of the experiments used for quantification out of a larger set of time lapses that were also analysed for statistical purposes. Diffusion analysis by raster image correlation spectroscopy (RICS) was analysed using SimFCS 4 software (G-SOFT Inc.) as previously described in.[45] Point spread function was determined as described elsewhere.[45] RICS images series (256 × 256 pixels) were taken using either an 8, or 4µs dwell time with no difference in the diffusion yielded between them. Each time lapse was taken for 200–300 total frames. From each full-frame time-lapse, a smaller region of interest was selected (32 × 32 pixels) and diffusion coefficient was obtained by fitting the experimental 2D autocorrelation function to a single diffusion mode. The 2D autocorrelation map was then fitted to obtain a surface map, employing the characterized waist value and the appropriate acquisition values for line time and pixel time.
Acute Brain Slices Experiments
500–1000 µm slices were prepared from adult animals (6 to 7 weeks old) with a McIllwain tissue chopper and incubated at 30 °C in artificial cerebral spinal fluid (ACSF; in × 10−3 m, NaCl 125, NaHCO3 25, KCl 2.5, NaH2PO4 1.25, glucose 25, CaCl2 2, MgCl2 1) oxygenated by a 95/5 (v/v) O2/CO2 gas mixture. Slices were metabolically labeled, in situ, with fusogenic liposome composed of DOPE/DOTAP/chol-N3 (1/1/0.1) in ACSF for 2 h. After washing, slices were subjected to Copper-free click chemistry labelling using 50 × 10−6 m cy3-DBCO for 3 h. After extensive washing to remove nonreacted probe, tissue was fixed in 4% paraformaldehyde for 25 min. As a final step, fixed brain tissue was embedded 24 h in a mounting media composed of 70% sorbitol in PBS buffer containing 0.01% sodium azide, which help to align the refractive index of the objectives and tissue confering higher-resolution and imaging depth. Samples were imaged by Leica Microsystems using a Leica TCS SP5 confocal microscope with the Leica HC Fluotar 10/0.3, HC Apochromat 20/0.77, and HC Apochromat 40/0.75. Image reconstructions were performed using Imaris Imaging software (BItplane). After imaging, samples were store in PBS containing 0.01% sodium azide at 4 °C.
Statistics
Data were analyzed using SPSS statistics software. Results of Kruskal Wallis test are indicated by: not significant (ns) (p > 0.05), * (p < 0.05), ** (p < 0.01), and ***(p < 0.001).
Acknowledgements
M.L., O.T., and J.A.N.-G. contributed equally to this work. This work was supported by grants from the Spanish Ministry of Science Innovation and Universities, (Grant No. BFU-2015-68981-P) and the Basque Government (Grant No. IT1264-19) to F.-X.C. and M.L.. The authors thank J. M. Gonzalez Mañas and Sergio Perez Acebron for helful comments on the manuscript. The authors thank the technical and human support provided by the analytical and high-resolution microscopy facility (SGIker) of UPV/EHU and European funding (ERDF and ESF). J.B.d.l.S. acknowledges funding from the Bill and Melinda Gates Foundation and the BBSRC (Grant Nos. INV-016631 and BB/V019791/1, respectively). This work was supported in part by the Fundación Biofísica Bizkaia (FBB) and the Basque Excellence Research Centre (BERC) program of the Basque Government. J.A.N.-G. was supported by a FI predoctoral fellowship from the Basque Government and currently by FBB.
Conflict of Interest
M.L. and F.-X.C are coinventors on a patent application filled by the University of the Basque Country (UPV/EHU) concerning the described technology.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.