The Internal Structure of the Velvet Worm Projectile Slime: A Small-Angle Scattering Study
Abstract
For prey capture and defense, velvet worms eject an adhesive slime which has been established as a model system for recyclable complex liquids. Triggered by mechanical agitation, the liquid bio-adhesive rapidly transitions into solid fibers. In order to understand this mechanoresponsive behavior, here, the nanostructural organization of slime components are studied using small-angle scattering with neutrons and X-rays. The scattering intensities are successfully described with a three-component model accounting for proteins of two dominant molecular weight fractions and nanoscale globules. In contrast to the previous assumption that high molecular weight proteins—the presumed building blocks of the fiber core—are contained in the nanoglobules, it is found that the majority of slime proteins exist freely in solution. Only less than 10% of the slime proteins are contained in the nanoglobules, necessitating a reassessment of their function in fiber formation. Comparing scattering data of slime re-hydrated with light and heavy water reveals that the majority of lipids in slime are contained in the nanoglobules with homogeneous distribution. Vibrating mechanical impact under exclusion of air neither leads to formation of fibers nor alters the bulk structure of slime significantly, suggesting that interfacial phenomena and directional shearing are required for fiber formation.
1 Introduction
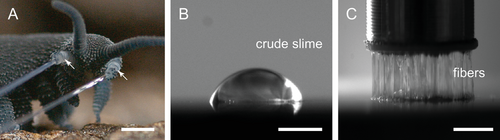
From a materials science and materials processing perspective, the ability of certain biological organisms such as spiders[1] and mussels[2] to rapidly produce high-performance polymeric materials from condensed protein solutions is highly relevant to current efforts to develop sustainable polymeric materials. Recently, the projectile slime of velvet worms (onychophorans) has emerged as an exciting model system for inspiring development of circular recyclable plastics and adhesives.[3] Velvet worms comprise an evolutionary old group of invertebrates which are distributed in tropical and temperate forests of the southern hemisphere. A common feature of all velvet worms species is that they use an adhesive slime to defend against predators and capture prey such as insects, woodlice, and other soil-dwelling invertebrates. The initially liquid slime is ejected from two specialized nozzles on either side of the velvet worm head (Figure 1A) by strong muscle contractions and spreads over the prey or opponent, which becomes more and more entangled in the forming adhesive threads as it tries to escape.[4-6] It has been proposed that the slime is stored in a condensed liquid phase consisting largely of a suspension of proteins (Figure 1B); yet, in mid-air, the slime transitions into a viscoelastic gel phase that can then be drawn into long sticky fibers that become stiff and glassy upon drying (Figure 1C). The dried fibers are dissolvable in water and new fibers can be drawn from the solution.
Analyses of the biochemical composition of the slime in various onychophoran species revealed that it contains ≈90% water. The dry mass is composed of ≈50% proteins, ≈2% carbohydrates, which are mainly linked to proteins, and ≲1% lipids. The remainder of the dry mass is mainly undescribed; however, it presumably consists of small solutes such as ions or free amino acids. Three molecular weight classes of proteins were found in different species, such as proline-rich high molecular weight (HMW) proteins, but also smaller proteins that are present at lower concentrations.[7-11] A first effort to identify slime proteins in Euperipatoides rowelli by matching expressed sequence tags to separated proteins revealed HMW proline-rich proteins, smaller concentrations of lectins and small, possibly antimicrobial, peptides.[7] A more recent study on a still non-described but most likely distantly related species of Eoperipatus (Peripatidae) from Singapore[11] was able to reconstruct the sequences of two hydroxyproline-containing HMW proteins of 230 and 190 kDa and a few lower-molecular-weight proteins by transcriptomic sequencing and proteomics. They found that the HMW proteins presumably together with lower MW cysteine-rich proteins build HMW multi-protein complexes linked by disulfide bonds. However, a complete identification of predominant proteins in velvet worm slime has not been achieved so far.
Previous nanostructural analysis of the slime with cryo-transmission electron microscopy (cryo-TEM), stimulated emission depletion (STED) microscopy, dynamic light scattering (DLS), and atomic force microscopy (AFM) revealed the presence of spherical nanoglobules with diameters on the order of 100 nm, which were found to consist of proteins and possibly lipids according to fluorescent staining combined with STED microscopy. Without experimental demonstration, it was assumed that the proteins responsible for building the fiber core are mainly stored in these nanoglobules, which limited the consideration of alternative functions of the nanoglobules.[3, 11] Evidence from vibrational spectroscopy and X-ray diffraction studies suggests that proteins in the slime exist, at least partially, in a β-crystalline conformation similar to that in spider silk, but that this structure is partially unfolded and lost following shear mechanical forces inherent in the fiber formation process.[12] Indeed, it was proposed that protein unfolding is a critical step in the activation process necessary for forming fibers, perhaps by preferring inter-molecular interactions between protein chains rather than intra-molecular, likely mediated via electrostatic linkages.[12] While the existence of β-crystallines were confirmed by Lu et al.,[11] they suggest that cysteine-based complexation and low-complexity domains in the N-termini of HMW proteins, which are known to favor liquid–liquid phase separation (LLPS), are the essential mechanisms for slime formation.[11] In spite of these key insights into the molecular formation mechanisms of velvet worm slime fibers, open questions have remained unanswered. In particular, the nanostructural organization of specific slime proteins and other biomolecules within the slime has not been quantitatively analyzed. This is critical to understand the underlying physical and chemical principles in order to transfer the reversible fiber-forming mechanism to synthetic polymers or to develop sustainable bio-inspired polymer processing strategies.
Here, we applied a combination of small angle neutron scattering (SANS) and small angle X-ray scattering (SAXS) to investigate the size and distribution of protein and lipid components within native and re-hydrated velvet worm slime of the peripatopsid species E. rowelli under resting conditions and under mechanically agitated conditions. Small-angle scattering with X-rays and neutrons is uniquely suited to investigate biological soft matter[13, 14] because it can access nanometric length scales, can provide truly sample-averaged structural information, and can be applied under physiologically relevant conditions. The combination of SAXS with SANS, together with contrast variation achieved by replacing regular water with D2O in the re-hydrated slime, enables the unambiguous differentiation between protein, lipid, and aqueous components of biomolecular assemblies. We find that only a small fraction of the proteins present in the slime are assembled into nanoglobules, while the majority exists as free proteins in the continuous liquid phase. Therefore, it might be reasonably assumed that HMW precursors of fiber cores are free in solution and not stored within nanoglobules, which is in contrast to the previous model of fiber formation in velvet worm slime. Although the overall lipid content in the slime is low, our findings indicate that they are localized predominantly within the nanoglobular fraction. Without the introduction of additional interfaces and directional shearing, mechanical agitation is found to have no significant influence on the nanoscale structural features in slime studied by SANS and SAXS, although it causes macroscopic gelification.
2 Results and Discussion
2.1 Determination of the Molecular Weight of Slime Proteins
In order to gain a deeper understanding of the rapid and reversible mechanoresponsive material behavior, we studied the nanostructural organization of the velvet worm slime of E. rowelli. Since proteins are the major macromolecular component, we analyzed the overall protein composition of the slime as basis for modeling of the SANS and SAXS data. Using denaturing polyacrylamide gel electrophoresis (SDS-PAGE), proteins were separated (Figure 2) and molecular weight of protein bands was calculated based on the migration distances in relation to the protein standard scale (Table S1, Supporting Information). Band intensities were plotted and the mass ratio of bands was calculated by comparing the areas under the intensity curves. Molecular weight fractions were formed by averaging the molecular weight of protein bands under consideration of their mass ratio (Supporting Information). Pre-assumptions for SANS and SAXS modeling are based on these averaged values of molecular weight fractions.
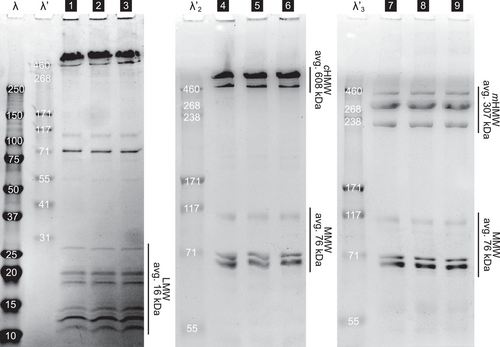
The predominant protein bands occur in three distinct fractions of molecular weight, which is in line with reports from previous studies.[7, 9] Under non-reducing conditions, two prominent bands of 634 and 478 kDa in a mass ratio of 5:1 appear in the HMW region of the gels (lanes 1–6, Figure 2). We average these bands to 608 kDa with the designation HMW complex mass , since they consist of monomers linked by disulfide bonds. Under reducing conditions they dissociate into lower molecular weight bands at 429, 323 and 232 kDa (mass ratio of 1:6:3) which averages to the HMW monomer mass = 307 kDa. Complexation of proline-rich monomers into HMW protein complexes was also reported in a recent study of a distantly related species, Eoperipatus sp., from Singapore.[11] There, the molecular weight of the predicted monomeric protein sequences was found to be 230 and 190 kDa assembling into HMW complexes in native slime[11] which further supports our findings. In E. rowelli, under native conditions the monomeric form of HMW proteins exists as weak bands in the SDS-gels (lane 1–3, Figure 2). In the ≈100 kDa range, three main monomeric bands (108, 70, and 69 kDa in a mass ratio of 1:1.5:3) occur which yield the average mid molecular weight (MMW) mass of mMMW = 76 kDa. In the region of low molecular weight (LMW) at least 10 protein bands appear between 11–27 kDa, with an average mass of mLMW = 16 kDa. The HMW, MMW, and LMW fractions appear in an overall mass ratio of 60:10:30 (detailed gel data in Supporting Information).
2.2 Nanoscale Structure of the Native Slime
| [vol%] | [%glob. vol.] | [%glob. vol.] |
|---|---|---|
| 4.5 ± 0.5 | ≈70 | ≈30 (25)a) |
| Globules | HMW proteins | LMW proteins | |
|---|---|---|---|
| ϕprot [vol%] | 0.4 ± 0.04 | 2.7 ± 0.5 | 1.4 ± 0.3 |
| =8 ± 1% | =60 ± 10% | =32 ± 10% | |
| R [nm] | 41 ± 2 | 6 (10)b) ± 1 | 1.5 ± 0.5 |
| δR [nm] | ≈6 | - | - |
| Exponent ν | - | 0.2 ± 0.05 | 0.2 ± 0.05 |
| Complex fraction [%] | - | 20 ± 10 | - |
- a) When accounting for 5 % lipids
- b) For complexes.
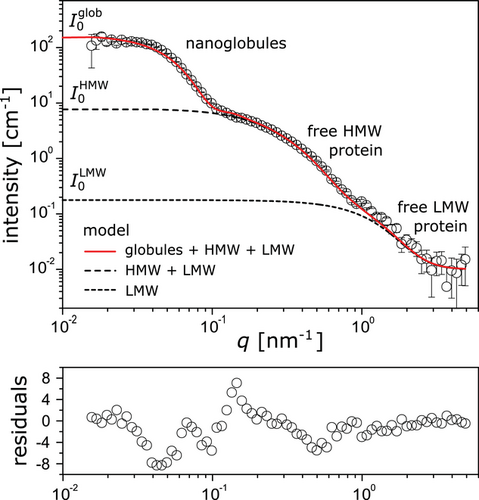
The experimental data (symbols in Figure 3) are well described by the model (solid line in Figure 3) based on this multi-population-approach upon adjustment of the model parameters. The best-matching parameter values yield the average size and polydispersity of the nanoglobules and the gyration radii and scaling exponents of the free proteins (Table 1). For the average globule radius we obtain Rglob = 41 ± 2 nm, with a moderate polydispersity of δRglob ≈ 6 nm. Previous DLS, AFM and cryo-TEM measurements revealed globule radii in a similar range, although slightly larger (DLS, hydrodynamic radius ≈75 nm). Small-angle scattering can, however, be considered to be the more reliable technique for radius determination of nanoglobules, since it provides representative, sample-averaged values whereas the hydrodynamic radius from DLS includes the collective motion of the sphere with its counter ion cloud or the sample preparation for AFM and cryo-TEM may skew the results. The best-matching gyration radii of the free proteins, obtained in a self-consistent parameter adjustment to the scattering curves of both the native slime and the re-hydrated slime discussed further below, are = 6 ± 1 nm, = 10 ± 1 nm, and RLMW = 1.5 ± 0.5 nm, respectively, where m and c denote again the monomeric and complexed forms. The fits yield very small scaling exponents, ν < 1/3, which suggests that the proteins adopt conformations of crumpled globular objects[19] as was previously found for single chain nanoparticles[20] and implies the presence of small compact regions in the proteins. Indeed, the slime proteins were reported to comprise compact β-sheet regions and less compact random coiled regions.[11, 12]
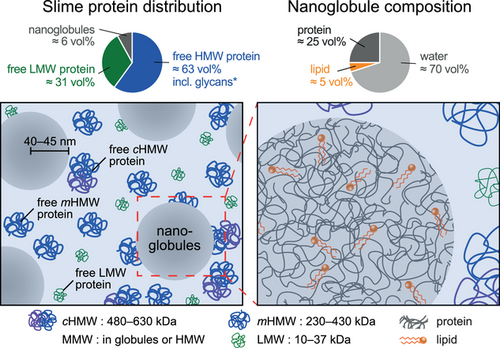
Importantly, in contrast to previous models and assumptions, only a small fraction of the protein volume is contained within the globules, while most of the proteins exist freely in the liquid slime. In the native slime, approximately one fifth of the HMW proteins are engaged in complexes, according to the model, which is in contrast to the high levels of HMW complexes suggested by SDS-PAGE under non-reducing conditions. Our finding that large quantities of protein exist outside of the nanoglobules is further supported by re-assessing previously published but differently interpreted data. A strong protein signal is clearly visible outside the nanoglobules in non-deconvoluted STED-micrographs published in Supporting Information of Baer et al. 2017, (Figure S5b, Supporting Information).[3] This result has important implications for the understanding of fiber formation in velvet worm slime. These SAS results clearly show that the continuous phase contains the main part of the proteins (≳90%) including the HMW proline-rich protein species, which are believed to be the precursors of fiber cores. We therefore conclude that the nanoglobules do not contain the bulk of solid material which forms the fiber core upon mechanical agitation which is in contrast to a previous suggestion.[3] As a consequence, this requires a re-consideration of the functions of nanoglobules in the realization of a stable storage state and during the transition from fluid into solid fibers induced by a specific mechanical trigger.[10, 12, 21] One may speculate that the nanoglobules serve as nuclei for protein aggregation, increasing the rate of material stiffening upon application of mechanical force.[22, 23] Alternatively, comparable to snail[24-27] or hagfish mucuses,[28, 29] during the fiber formation process in velvet worm slime, nanoglobules might release a cross-linking agent due to rupture that might be involved in the versatile adhesion or in the material stiffening of the fiber core. In particular, an optimized adhesion of onychophoran slime to wax and lipoproteins in the cuticle of their prey would be achieved by nanoglobules which contain lipids and proteins with hydrophobic functions. However, further research is required to finally clarify the function of the nanoglobules.
We were furthermore able to determine the protein content in the slime from the scattering intensity on a quantitative level by accounting for the absolute SLD difference in Equation (2), and an overall protein volume fraction of = 4.5 ± 0.5% is obtained, where the uncertainty (0.5%) mainly reflects uncertainties in the average molecular mass and the degree of complexation of free HMW proteins. Given the typical protein mass density of 1.4 g cm−3,[15, 30] this value translates into a protein mass content of 6.3 ± 0.7 mass%, which is in acceptable agreement with the value reported in the literature (=4.7–5.5 mass%).[8, 10] In fact, this agreement appears to be an independent validation of the model assumptions that were made. This result is also essentially independent from the exact value of because is largely dominated by the free proteins.
It should be noted that the SANS features at higher q must arise from free proteins rather than the globules’ internal structure. To illustrate this point, let us assume that all protein mass would be contained in the nanoglobules and the other two scattering features would only represent substructures of the globules. Following Figure 2, with a protein volume fraction of ≈4%, the SLDs noted in Table 2, a volume of the globules of 270 000 nm3 and no additional solvent in the globules, I0 ≈ 6500 cm−1 would result, which is clearly inconsistent with the experimental data in Figure 3. On the other hand, if we added solvent to the globules such that their volume fraction would be given by ϕglob = ϕprot + ϕsolv and their SLD by , the associated intensity would be reduced, but even in the non-physical limit that all water is contained in the globules, ϕsolv = 1 − ϕprot, the resulting intensity (I0 ≈ 260 cm−1) would still be significantly above the experimentally observed value.
Overall, the uncertainties of the model parameters given in the tables are vastly dominated by systematic uncertainties introduced by the model assumptions and, as pointed out in earlier work,[31] much larger than the statistical uncertainties that can be quantified in a standard procedure via χ2 analysis.[32] The most critical assumptions are the average masses of the free proteins responsible for the mid-q and high-q features. For example, slightly different values for the volume fraction of free HMW proteins in solution are obtained when assuming they are all complexed and when coexistence of monomeric and complexed forms is assumed. Analogous reasoning applies to the volume fraction of free LMW proteins. Moreover, we were able to exclude the influence of temperature on the measurements. A temperature change from 20 to 4 °C leads to an increased viscosity of slime; however, it has no significant influence on the SANS curve and thus on the slime structure (Figure S1, Supporting Information).
2.3 Protein/Lipid Distribution
SANS measurements on the native slime are insensitive to the lipid distribution because of the similar SLDs of H2O and the lipid tails. However, considerable SLD contrast between water and lipids in SANS is generated when replacing H2O with D2O (isotopic contrast variation) and additional valuable information is obtained in complementary SAXS experiments. For the contrast variation the slime had to be treated by drying and subsequent re-hydration with D2O or H2O (see Experimental Section). In previous studies re-hydrated slime was reported to behave very similarly to native slime.[12] Nevertheless, native and re-hydrated slime must not be considered a priori to have identical structures. The characteristics of the re-hydrated slime are therefore summarized in a separate table (Table 3).
| [vol%] | [%glob. vol.] | |||||
|---|---|---|---|---|---|---|
| 3.1 ± 0.5 | 3.05 ± 0.5 | ≈0.05 | ≈70 | ≈25 | 5 ± 1 | |
| Globules | HMW proteins | LMW proteins | |
|---|---|---|---|
| ϕprot [vol%] | 0.20 ± 0.05 | 1.9 ± 0.3 | 0.96 ± 0.15 |
| =6 ± 1% | =63 ± 10% | =31 ± 10% | |
| R [nm] | 44 ± 2 | 6 (11)a) ± 1 | 1.5 ± 0.5 |
| δR [nm] | ≈6 | – | – |
| Exponent ν | – | 0.2 ± 0.05 | 0.2 ± 0.05 |
| Complex fraction [%] | – | 40 ± 15 | – |
- a) For complexes.
SANS curves of native slime and H2O-based re-hydrated slime are almost identical (Figure S2A, Supporting Information) up to a scale factor that reflects incomplete re-hydration. Indeed, according to visual inspection, a small part of the material remained undissolved, resulting in a lower material concentration, 3.1 ± 0.5 vol% for the H2O-based re-hydrated slime. The practically identical shapes of the curves, however, demonstrate that incomplete dissolution otherwise did not change the slime composition significantly. Comparing SAXS curves obtained with H2O-based and D2O-based re-hydrated slime (Figure S2B, Supporting Information) is meaningful because the X-ray SLD contrast only arises from the electron density difference, such that the SAXS curves are unaffected by isotopic contrast variation. Also these curves are essentially identical up to a scale factor (fHD = 1.30, used again later on) that merely reflects variations in the degree of re-dispersion. In other words, the replacement of H2O by D2O leaves the slime structure largely unaffected.
Comparing the small-angle scattering curves of the re-hydrated slime in SANS (namely the two isotopic contrasts H2O and D2O) and the electron density contrast in SAXS reveals a similar overall shape with distinct features associated with globules and free proteins (Figure 4). Most notably, the globule-related features are at exactly the same q-values in all three contrasts, suggesting that scattering contrast arises mainly from the objects’ outer shape. In other words, the composition of the globules can be considered approximately homogeneous. The total protein content determined by SANS for the H2O-based re-hydrated slime, together with the electron density of dry protein = 12.2 × 10−6 Å−2[15] for reason of self-consistency sets the absolute scale of the SAXS data, for which no absolute calibration was available. The overall SANS intensity ratio between the H2O and D2O samples (defined via the respective values of ) is not quite as high as predicted by Equation (3) when assuming ρwat = ρ(D2O) and accounting for the concentration difference. The discrepancy must be attributed to the fact that the D2O-based re-hydrated slime contains residual H2O due to incomplete drying at ambient humidity and due to exchange with labile hydrogen atoms of the non-aqueous material. The best-matching water SLD, ρwat = 5.1 × 10−6 Å−2, coincides with 15% of H2O remaining, which appears to be plausible. The distinct intensity upturn in the low-q-limit (seen best in the D2O sample) indicates the presence of larger aggregates which are known to contribute to the scattering intensity at low q considerably even at very low concentrations.[33, 34] The aggregates are likely the result of imperfect re-dispersion of the slime after drying and are therefore not visible in the scattering curves from native slime. One may speculate that in the D2O sample they occur more because proteins are more prone to aggregation[35] or they are better seen because of less efficient sedimentation of aggregates in the D2O.
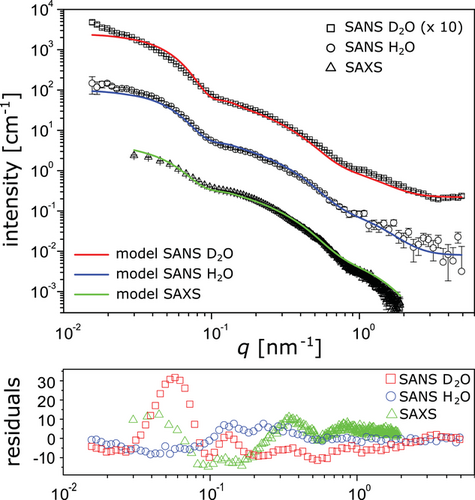
A small but noticeable difference between the SAS curves from native and re-hydrated slime is that in the latter case the feature associated with the free HMW proteins is somewhat flattened. This observation suggests the presence of two sub-populations with comparable size, which we attribute to monomeric and complexed forms of HMW proteins. Self-consistent modeling of all data sets from the re-hydrated slime is found to work best when a complex fraction of 40 ± 15% is assumed. The significantly less flattened shape of the corresponding feature in the native slime, on the other hand, suggests that one population dominates. Indeed, we obtain a complex fraction of only 20 ± 10% when imposing consistent structural parameters of monomeric and complexed HMW proteins also for the native slime SANS analysis. Different complex fractions in native and re-hydrated slime suggest that re-hydration does not leave the slime structure entirely unaffected but likely leads to variations in characteristics that are very sensitive to the experimental conditions.
A closer inspection of the mostly similar shape of the three scattering curves obtained with re-hydrated slime reveals an important difference: The intensity ratio in the scattering by the globules and by the free proteins (encoded for example in the ratio ), is significantly different for the SANS H2O, SANS D2O, and SAXS contrasts, indicating that nanoglobules and free proteins have a different non-aqueous composition. Within the framework of the model this must be attributed to the presence of lipids in the globules, as previously proposed.[3] Even though the overall lipid content in the slime dry mass is low (≲1 mass%),[8] the lipids are sufficiently abundant to constitute a significant volume fraction of the globules, because the latter only contain a small fraction of the total protein mass (see previous subsection). While the lipids’ hydrocarbon chains essentially do not contribute to the scattering in the H2O SANS contrast (, see previous subsection), in the D2O SANS contrast they enhance the scattering of the globules (). Conversely, hydrocarbon chains with their low electron density reduce the scattering of the nanoglobules in the SAXS contrast, because they partially compensate the higher electron density of proteins (). As a consequence, the ratio is highest for the D2O SANS contrast and lowest for the SAXS contrast, the H2O SANS contrast being intermediate.
A finite volume fraction of lipids in the nanoglobules (Table 3) is thus taken into account by the simulated intensities according to the best-matching parameters in the common model (solid lines in Figure 4). The best simultaneous agreement with all scattering contrasts is achieved for 5%, corresponding to an overall lipid content of ≈1.5 vol% in the macromolecular dry mass or ≈0.8 mass% in the total dry mass (assuming a protein density of 1.4 g cm−3[15, 30] and an alkyl chain density of 0.75 g cm−3), in line with the ≲1 mass% reported earlier.[8] This result suggests that most or even all lipids are contained in the globules. If the nanoglobules take up most or all lipids but only a small fraction of the available proteins, then this could in turn indicate that nanoglobule formation may be limited by the availability of lipids. On the other hand, if the nanoglobules are mainly composed of MMW proteins, then those proteins could alternatively be the limiting factor for globule formation. These hypotheses can be tested in future studies.
A common model with zero lipid content in the nanoglobules leads to a poor agreement with the experimental data (Figure S3, Supporting Information). Overall, the common model, which describes the nanoglobules as homogeneous spheres, is in satisfactory agreement with all three small-angle scattering data sets. This result indicates that the nanoglobules do not exhibit any coarse-scale heterogeneity in their composition and rules out lipid/protein, core/shell architectures, which refines the model suggested previously by STED-microscopic investigations of the slime in Baer et al. 2017, Figure S5, Supporting Information.[3] In addition, since the amount of lipid in the nanoglobules (≈5 vol%) is insufficient to form a potential lipid layer on the globules, we suggest that the role of the lipids is to help agglutinate the proteins, influencing both size and relative monodispersity of nanoglobules in the velvet worm slime. This function can possibly be achieved through interactions between lipids in form of fatty acids and cationic residues or hydrophobic sites of nanoglobule proteins. In order to verify this hypothesis, further sequence data of the proteins and lipid/detergent treatment before slime re-hydration are required. Current knowledge on the chemistry of the slime lipids and acyl-chain base detergents is, however, still limited.[8]
2.4 Effects of Mechanical Agitation
In order to investigate nanostructural changes during mechanical agitation, SANS curves of native slime before and after vortexing and sonication were obtained (Figure 5 and Experimental Section). We focused on mechanical agitation without significant air interface by measuring completely filled containers in order to exclude interfacial effects. Mechanical treatment by vortexing leads to macroscopic gelification of the slime in the fully filled measurement cuvettes, however formation of fibers was not observed (Figure S4, Supporting Information). Nonetheless, the SANS curve of the vortexed slime (Figure 5, open triangles) remains virtually identical to the SANS curve of the untreated native slime (Figure 5, open circles). Sonication is a somewhat harsher treatment, immediately leading to gelification and rigidification of the slime, although again fiber formation was not observed in the completely filled cuvette (Figure S4, Supporting Information). Yet, there are still hardly any changes to the SANS curves (Figure 5, open diamonds). The only changes concern the low-q region, but we refrain from ascribing them to structural changes of the slime, because we suspect that they are rather due to the presence of sub-µm air bubbles generated during sonication.
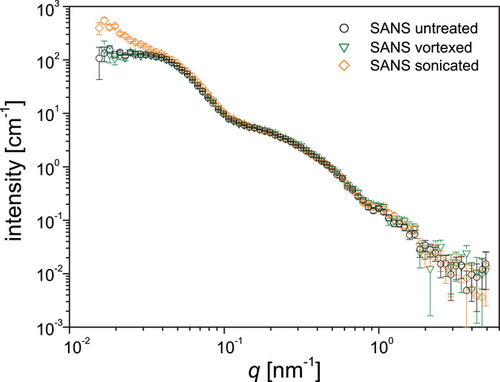
These results clearly demonstrate that macroscopic gelification occurs without any significant changes of the slime structure on the nanoscale level which indicates that nanoglobules stay intact after applying mechanical treatment, provided that sample preparation occurs within a fully filled container. Since fiber formation was not achieved under these conditions, we assume that directional shear at larger scales and the introduction of air or hydrophobic interfaces may be required to trigger the protein assembly into fibers. This is also consistent with previous measurements using FTIR spectroscopy coupled with in situ rheology[12] indicating that conformational changes in the backbone of slime proteins required fiber formation in presence of air, but could not be seen by mechanical shear alone. Although previous AFM studies of vortexed slime were interpreted as showing the disassembly of nanoglobules to form fibers, the current findings demand a reinterpretation of this model. Indeed, our findings indicate that 1) the majority of slime fiber proteins are not contained within the nanoglobules and 2) the nanoglobules are not ruptured during vortexing or sonication. Based on these observations, we conclude that the nanoglobules are not the source of fiber forming proteins and thus, their role in the slime must be reconsidered.
3 Conclusions
Small-angle scattering with neutrons and X-rays was utilized to investigate the bulk structure of the projectile slime of velvet worms under native conditions, after mechanical agitation, and after reconstitution by drying and re-hydration. The measurements revealed that only ≈6% of the slime's proteins is contained in the nanoglobules, while the rest exists in the form of free proteins found as monomers or HMW complexes in solution. The nanoglobules are spherical and show an average radius of ≈40–45 nm with rather narrow size distribution. In addition to a water content of ≈70 vol%, nanoglobules are comprised of ≈25 vol% proteins and a non-negligible lipid content of ≈5 vol% which is consistent with the reported overall low lipid content in the slime's dry mass suggesting that the major part of available lipids are in the nanoglobules. There is no indication that the nanoglobules exhibit any pronounced protein/lipid core/shell structure. Re-hydration or cooling does not significantly alter the structure of the slime as determined by scattering. Importantly, macroscopic gelification via mechanical agitation within fully filled containers leaves the slime structure largely unaffected. On the other hand, fiber formation may be caused by directional drawing and interfacial phenomena induced by the exposure of slime to hydrophobic interfaces such as air. This hypothesis will be tested in the future with the help of X-ray and neutron reflectometry.
In contrast to previous assumptions,[3, 11, 12] our data clearly show that HMW fiber-forming proteins freely exist in the solution, while only a small amount of protein and an even smaller amount of lipids form the nanoglobules (Figure 6). Recent studies on a Peripatidae species found in Singapore showed that specific low complexity sequences in the HMW proteins can be induced to undergo liquid–liquid phase separation, which was also suggested to play a role in nanoglobule formation.[11, 12] However, given the evolutionary distance between this species and E. rowelli (diverged over 380 MYA), it is not clear that a similar mechanism would be at play here. The previous model which assumed the nanoglobules to be the storage units of the fiber precursors,[3, 11, 12] thus needs to be reconsidered. With this result, the question arises again as to what function the nanoglobules fulfill in realizing a stable storage phase and during the mechanoresponsive gelification and fiber formation. It is conceivable that the nanoglobules serve for nucleation or release molecules which contribute to fiber stiffening and the adhesion on both hydrophilic and hydrophobic substrates. Future studies are required to assess the validity of these propositions. The identification of the dominant proteins in the nanoglobules, which possibly are within the MMW fraction, and the function of lipids will be an important step. This might be achieved by protein sequencing, immunolabeling with antibodies and detection via super resolution microscopy, lipid/detergent treatment before slime re-hydration, or SANS measurements on reconstituted slime in combination with selective deuteration.
4 Experimental Section
Specimens and Slime Collection
Experiments were performed on the peripatopsid species Euperipatoides rowelli.[36] Specimens of E. rowelli were obtained from decaying wood at the corresponding localities and maintained in the laboratory as described previously.[37] The animals were collected and exported under the following permit numbers: SL101720/2016, issued by NSW National Parks and Wildlife Service (Australia), and PWS2016-AU-001023, issued by Department of Sustainability, Environment, Water, Population and Communities (Australia). All animal treatments complied with the principles of laboratory animal care and the German law on the protection of animals. Slime samples for each experiment were obtained from several specimens by stimulating them to eject the slime into 500 µL Eppendorf tubes. Collected slime was stored in the fridge for no longer than 4 days at 4 °C to avoid bacterial growth or potential degradation of proteins.
Chemicals and Sample Preparation
Unless stated otherwise, chemicals were purchased from Sigma (St. Louis, MO, USA) and used as received. Water: MilliQ water (H2O, MilliQ Integral ultrapure water Type 1, specific resistance ≥18.2 MΩm, organic content ≤5 ppb). Right after collection, slime samples were treated with NaN3 (0.01%) in order to avoid bacterial growth. Re-hydrated samples in H2O and D2O were prepared by drying 200 µL of slime for 4 h at ambient air and nitrogen flow. The 20 mg of dried material was resuspended in 200 µL of H2O or D2O, respectively, by smoothly shaking at 30 °C at an orbital thermoshaker (Biosan TS-100). Native and re-hydrated slime were slowly (≈1 min) pipetted into cuvettes (Type 32, Fernes UV Quarz, 1 mm path length, Starna Scientific Ltd.) using manual syringes (22ga needle with 0.72 mm orifice) without causing noticeable gelification or fiber formation. In case of experiments on non-agitated slime, samples were incubated for 30 min at ambient conditions for material relaxation in order to avoid any possible aggregations due to pipetting. Vortexing was performed by shaking slime filled cuvettes 10 s at 1000 rpm. Sonication was applied to slime filled cuvettes in a ultrasonic bath for 10 s at 40 kHz (Figure S4, Supporting Information).
Denaturing Polyacrylamide Gel Electrophoresis (SDS-PAGE) of Slime Proteins
The distribution of molecular weight of slime proteins was determined using denaturing polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 2). The full range of protein bands was obtained with a 4–20% gradient polyacrylamide Mini-PROTEAN TGX Precast SDS gels (Bio-Rad, Montréal, Canada), using Mini-PROTEAN II electrophoresis cell (Bio-Rad, Montréal, Canada). High resolution in the HMW region was achieved by using hand-cast 5% polyacrylamide gels (4.6 mL H2O, 2 mL 1.5 m Tris-HCl, pH 8.8 buffer, 80 µL SDS 10%, 1.33 mL 30% acrylamide:bis-acrylamide 29:1, 20 µL ammonium persulfate (APS) 20%, 5 µL TEMED). For sample preparation, crude slime was diluted ten times in water. A range of concentration and thermal treatments were tested beforehand (data not shown). The optimal treatment to describe native slime under non-reducing conditions while preventing protein to aggregate and be stuck on top of the SDS-page gel was a final slime diluted 100 times with SDS sample buffer according to Laemmli[38] followed by a thermal treatment at 65 °C for 5 min. For SDS-PAGE under reducing conditions, 5% β-mercaptoethanol was added prior to the same thermal treatment. 10 µL of sample was injected per well and tris-glycine SDS running buffer (25 mm Tris, 192 mm glycine, 0.1% SDS, pH 8.3) was used at 10 mA for 15 min then 50 mA for ≈2 h. The prestained protein ladders (Precision Plus Protein Kaleidoscope, Bio-Rad, Montreal, Canada) in the molecular range 10–250 kDa and the HMW protein ladder HiMark (Thermo Fisher Scientific, Montréal, Canada) in the molecular range 31–460 kDa were used. After electrophoresis, total protein staining was performed using Coomassie Blue (0.05 %w/v Coomassie Blue R-250) for 40 min and destaining (30 % v/v methanol and 10 %v/v glacial acetic acid) for ≈5 h. Gels were documented using ChemiDoc MP and ImageLab (Bio-Rad, Montréal, Canada). Final image editing and panel design were performed using Adobe (San Jose, CA, USA) Photoshop CS5 and Illustrator 2020.
SANS Experiments
SANS was carried out on the Sans2d small-angle diffractometer at the ISIS Pulsed Neutron & Muon Source (STFC Rutherford Appleton Laboratory, Didcot, U.K., http://www.isis.stfc.ac.uk).[39] A usable q-range of 0.015–5 nm−1 was achieved utilizing an incident wavelength range of 1.75–12.5 Å and employing two detectors at 5 and 12 m from the sample. Each raw scattering data set was corrected for the detector efficiencies, sample transmission, and background scattering from the empty cell and converted to absolute scale with a standard sample (a solid blend of hydrogenous and perdeuterated polystyrene) using the software Mantid (http://www.mantidproject.org). The first test SANS measurement on native slime was conducted at the SANS-1 instrument operated by Hereon and FRM II at the Heinz Maier–Leibnitz Zentrum (MLZ), Garching, Germany.[40] The obtained data are shown in Figure S5, Supporting Information.
SAXS Experiments
SAXS measurements were done at the Materials Characterization Laboratory of the ISIS facility on a Nano-inXider instrument (Xenocs, Sassenage, France) using a micro-focus sealed-tube Cu 30W/30 µm X-ray source. The scattered X-rays were detected using a Dectris Pilatus 3 hybrid photon counting detector at a distance of 938 mm from the sample stage, and covering a usable q range of 0.03–2 nm−1. Scattering from the samples was collected in 1-mm borosilicate glass capillaries at room temperature. Data reduction (azimuthal averaging, buffer subtraction, absolute scaling) was done using the Foxtrot software.[41]
Data Analysis
Acknowledgements
The authors thank the ISIS Facility for the provision of beamtime on Sans2d (DOI: 10.5286/ISIS.E.RB181 0607) and the use of the Materials Characterization Laboratory, MLZ for allocation of a test beamtime, S. Busch for performing first SANS measurements, I. Marcotte for laboratory and technical support, A. A. Arnold for thoughtful discussion and inputs, and D. M. Rowell, I. S. Oliveira, C. Martin, and I. Schumann for help with specimen collection. A.B. thanks Robert J. Kittel for providing office and laboratory space in Leipzig. Collecting and export permits have been kindly provided by the Office of Environment & Heritage (NSW National Parks & Wildlife Service) and the Department of the Environment of Australia for the Australian species studied, and by the National System of Conservation Areas (SINAC, MINAE). A.P. thanks the Fonds de Recherche du Québec-Nature et Technologies (FRQNT), as well as the Quebec Network for Research on Protein Function, Engineering, and Applications (PROTEO) for the award of scholarships. A.B., G.M., and S.S. gratefully acknowledge support from the German Research Foundation (DFG: MA 4147/7-1 and 4147/7-2 to G.M.; SCHM 2748/5-1 and SFB 1208, project Z02, to S.S.). M.J.H. acknowledges support from the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant RGPIN-2018-05243). E.S. gratefully acknowledges financial support by the Max Planck Society and by the DFG via Emmy-Noether grant SCHN 1396/1.
Open access funding enabled and organized by Projekt DEAL.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
A.B., E.S., and I.H. designed the research; A.B. collected and prepared slime samples; N.M. and A.B. performed the experiments; E.S., I.H., and N.M. analysed the data. A.P. and A.B. performed and analyzed SDS-PAGE. A.B., E.S., and A.P. designed the figures; A.B. and E.S. wrote the manuscript; all authors discussed the results and approved the final manuscript.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.




