Genomic Signatures Correlating With Adverse Pathologic Features in Men Eligible for Active Surveillance
Jamie Michael and Eric V. Li are Co-first authors
ABSTRACT
Background
Genomic biomarkers offer opportunities to improve risk stratification for patients with prostate cancer. We performed a transcriptomic profile of active surveillance (AS)-eligible patients who underwent radical prostatectomy (RP) to understand genomic associations with adverse pathologic features (APF) at RP.
Methods
Patients considered AS-eligible (NCCN low-favorable intermediate risk) but proceeded to RP from February 2012 to September 2024 were identified from our prospective institutional database. Outcomes were classified by presence or absence of APF at RP, which was defined as grade group (GG) ≥ 3, pT3b, or pN1 disease. Previously established genomic signatures of interest were compared between the two groups. Scaled multivariable logistic regression was performed to evaluate associations between multiple genomic classification systems and the outcome of APF.
Results
There were 184 AS-eligible patients, of whom 153 (83.2%) had favorable intermediate risk disease and 31 (16.8%) had low risk disease. There were 41 patients (22.3%) who had APF at RP. The incidence of favorable intermediate risk disease did not differ between those with and without APF (80.5% vs. 83.9%, p = 0.64). Patients with APF had a higher baseline PSA (5.6 ng/mL vs. 4.9 ng/mL, p = 0.01) and Decipher score (0.55 vs. 0.41, p = 0.004) compared to those without APF. On scaled logistic regression with adjustment for log-transformed PSA, the Decipher score, PTEN loss, activated CD4, and ERG positive rate were significantly associated with APF (OR 1.61, 95% CI 1.11–2.32, p = 0.01). Of ten other previously published genomic classifiers, nine were significantly associated with APF.
Conclusion
AS-eligible patients with APF at RP demonstrate differences in gene expression when compared to those without APF. We established that multiple existing genomic classifiers not previously studied in this context demonstrate the ability to predict APF in this patient population. Inclusion of genomics in the risk stratification of AS-eligible patients has the potential to better inform clinical decisions.
1 Introduction
Prostate cancer is the most common solid organ malignancy in men, affecting 1 in 9 men [1]. While it remains the second leading cause of cancer death, prostate cancer varies widely in aggressiveness. Many cases of localized prostate cancer are indolent and do not necessitate immediate treatment. Active surveillance (AS) is preferred for National Comprehensive Cancer Network (NCCN) very low to low risk prostate cancer and may be considered for select favorable intermediate risk prostate cancer patients [2]. Despite this, over half of men placed on surveillance will go on to treatment, and more importantly, about 15% of men put on surveillance will have “extreme” reclassification to grade group (GG) 3 and above [3-6].
Traditionally, risk of adverse outcomes has been determined by clinical and pathologic features, such as prostate-specific antigen (PSA), grade group, or clinical stage. However, previous studies have shown a weak association between these pathologic variables and adverse pathologic features at time of surgery [7]. Molecular profiling has emerged as a biomarker to further differentiate men who may be at risk of adverse outcomes. Previous studies have demonstrated that the commercially-available Decipher Prostate Genomic Classifier (Veracyte Inc., San Diego, CA) is positively correlated with progression or presence of unfavorable pathology in patients considering AS [8-11]. There are, however, other genomic signatures which may demonstrate utility as well. For example, the prediction analysis of microarray 50 (PAM50) system, originally developed to classify breast cancer based on molecular subtypes, has been adapted for use in prostate cancer [12, 13]. More recently, the development of a prostate subtyping classifier (PSC) has identified four subtypes with distinct biological and clinical features [14]. While these classification tools have been shown to potentially aid in treatment decisions for men with locally advanced or metastatic prostate cancer, they have not been investigated in patients who may be candidates for AS.
As such, we performed a comprehensive transcriptomic profile of AS-eligible patients who underwent radical prostatectomy (RP) to understand which genomic biomarkers and subclassifications are associated with adverse pathologic features (APF) at RP. We also sought to compare association of genomic signatures with APF in the context of clinical and pathologic variables.
2 Methods
2.1 Patient Cohort and Clinical Characteristics
We performed a retrospective review of patients who would be eligible for AS but proceeded to RP at Northwestern Memorial Hospital from February 2012 to September 2024. This study was approved by the Institutional Review Board at Northwestern University (STU00214996). The study utilized retrospective deidentified data and patient consent as not required. The study was performed in accordance with the Declaration of Helsinki.
AS-eligible patients were defined as patients with NCCN low risk (GG 1, clinical stage T1c-T2a, and PSA < 10 ng/mL) or favorable intermediate risk prostate cancer (GG 1-2 and < 50% positive cores with no more than one of the following criteria: PSA 10–20 ng/mL or clinical stage T2b-T2c). Patients who were on AS and underwent reclassification to a higher grade or NCCN risk group were included if the patient remained eligible for AS in the new risk group (i.e. NCCN low risk reclassifying to NCCN favorable intermediate risk). Prostate biopsy and RP specimens were examined by experienced genitourinary pathologists. Two cohorts were created based on the presence or absence of APF at RP, which was defined as GG ≥ 3, pathologic stage ≥T3b, or nodal involvement (pN1).
2.2 Decipher Testing and Transcriptomic Analysis
The prostate needle biopsy core with the highest grade and percentage of cancer involvement was sampled for genomic testing using the Decipher prostate genomic classifier (Veracyte, San Diego, CA). Patients in whom the biopsy tissue was not available were included by performing genomic testing on the RP specimen to extrapolate the genomic signatures that would have been present at biopsy.
The Decipher Genomics Resource for Intelligent Discovery (GRID) registry (NCT02609269) collects whole transcriptome tumor profiles for genomic discoveries, including a compendium of over 400 curated gene expression signatures (accessed September 7, 2024) for patients treated at our institution. We extracted gene expression levels for a wide array of adverse molecular features, including androgen receptor activity (AR-A), homologous repair (HR) deficiency, PTEN loss, RB loss, tp53 mutation, ERG over expression, and immune activity gene expression signatures [15-23]. The basal-luminal PAM50 and PSC subtyping models were included as well.
2.3 Statistical Analysis
Comparative statistics (Mann–Whitney U, Kruskal–Wallis, Chi-squared, Fisher's exact) were used to compare clinicopathologic and genomic characteristics between patients with and without evidence of APF at RP. To further understand the potential for genomic profiling to predict APF in the context of clinical variables, we performed scaled multivariable regressions of ten commonly referenced studies and generated odds ratios per standard deviation increase in the biomarker [24-33]. All analyzes were performed in R Statistical Software v4.2.1 with statistical significance set at α = 0.05.
3 Results
We identified 184 AS-eligible patients who underwent RP. There were 153 (83.2%) with NCCN favorable intermediate risk prostate cancer and 31 (16.8%) with NCCN low risk disease. Within our cohort, 41 (22.3%) demonstrated APF at RP, which was determined by the presence of GG ≥ 3 in 40 patients, in addition to pT3b disease in 4 patients. No patients had pN1 disease. Patients with APF did not demonstrate any differences in age, race, BMI, comorbidities, prostate volume, or PSA density when compared to patients without APF (Supplemental Table S1). However, patients with APF did have a higher median PSA (5.6 vs. 4.9 ng/ml, p = 0.01) compared to those without APF. There was no significant difference in NCCN risk distribution between the two groups (p = 0.64) (Supplemental Table S1).
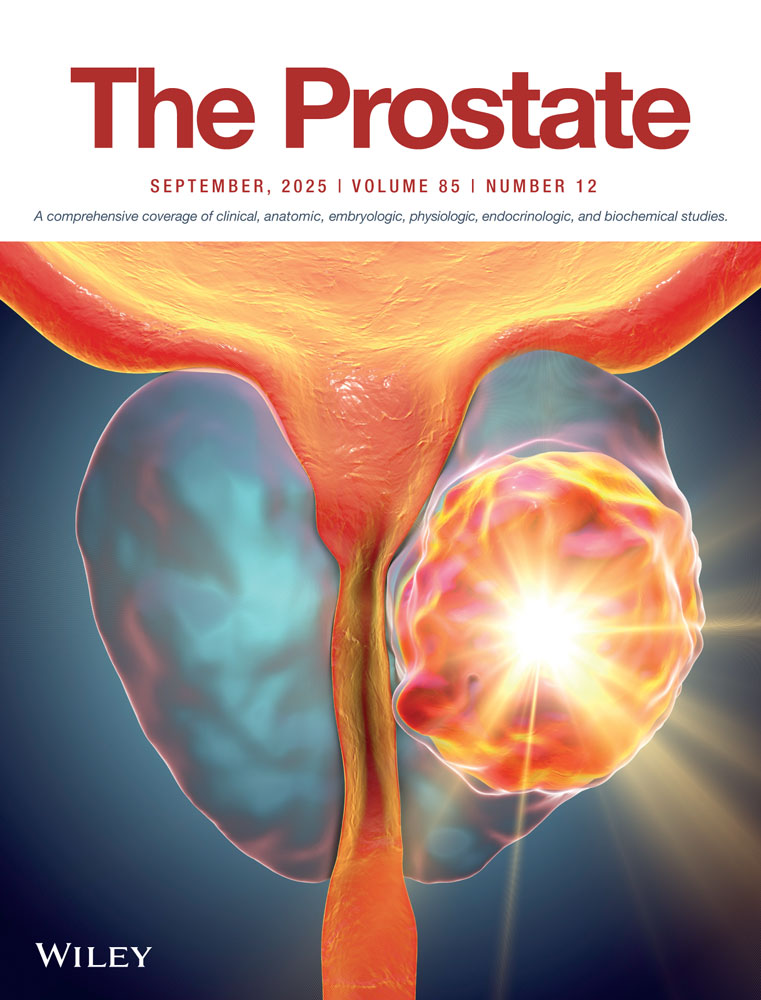
First, we sought to establish association between various genomic signatures and classifiers with the APF endpoint. Patients with APF demonstrated higher median Decipher scores (0.55 vs. 0.41, p = 0.004) (Table 1). Patients in the APF group were more likely to be ERG positive (65.9% vs. 44.1%, p = 0.02), have higher scores for a PTEN tumor suppressor loss gene expression signature (0.09 vs. 0.01, p < 0.001), and have higher activated CD4 expression (−0.02 vs. −0.07, p < 0.001) than patients in the non-APF group. RB deficiency and tp53 mutation signature scores were low and there were no differences between the two groups. With respect to PAM50 classification, the APF group had a lower proportion of the basal subtype (17.1% vs. 34.3%, p = 0.03) and higher proportion of luminal B subtype (41% vs 22%, p = 0.03) (Figure 1, Table 1).
| Adverse pathological features | |||
|---|---|---|---|
| Genomic characteristics | Absent (n = 143, 78%) | Present (n = 41, 22%) | p value |
| Decipher score, median (Q1, Q3) | 0.41 (0.26, 0.60) | 0.55 (0.38, 0.75) | 0.004 |
| Decipher risk, n (%) | |||
| Low | 77 (54%) | 14 (34%) | 0.03 |
| Intermediate | 30 (21%) | 8 (20%) | |
| High | 36 (25%) | 19 (46%) | |
| AR activity, median (Q1, Q3) | 13.4 (12.4, 14.2) | 13.6 (12.9, 14.7) | 0.09 |
| AR activity category, n (%) | |||
| Higher AR-A | 122 (85%) | 39 (95%) | 0.11 |
| Lower AR-A | 21 (15%) | 2 (5%) | |
| HR deficiency score, median (Q1, Q3) | −0.19 (−0.22, −0.16) | −0.18 (−0.22, −0.14) | 0.40 |
| HR status, n (%) | |||
| Intact | 98 (69%) | 24 (59%) | 0.26 |
| Deficient | 45 (31%) | 17 (41%) | |
| RB status, n (%) | |||
| Intact | 141 (99%) | 40 (98%) | 0.53 |
| Deficient | 2 (1%) | 1 (2%) | |
| tp53 mutation score, median (Q1, Q3) | 0.13 (0.06, 0.24) | 0.16 (0.06, 0.26) | 0.56 |
| tp53 status, n (%) | |||
| Wildtype | 133 (93%) | 37 (90%) | 0.51 |
| Mutant | 10 (7%) | 4 (10%) | |
| PAM50 subtype, n (%) | |||
| LuminalA | 62 (43%) | 17 (41%) | 0.03 |
| LuminalB | 32 (22%) | 17 (41%) | |
| Basal | 49 (34%) | 7 (17%) | |
| PSC subtype, n (%) | |||
| Luminal differentiated | 61 (43%) | 12 (29%) | 0.14 |
| Luminal proliferating | 14 (10%) | 9 (22%) | |
| Basal immune | 56 (39%) | 18 (44%) | |
| Basal neuroendocrine | 12 (8%) | 2 (5%) | |
| ERG subtype, n (%) | |||
| ERG negative | 80 (56%) | 14 (34%) | 0.02 |
| ERG positive | 63 (44%) | 27 (66%) | |
| pTEN loss score, median (Q1, Q3) | 0.01 (0.00, 0.04) | 0.09 (0.01, 0.38) | < 0.001 |
| pTEN loss status, n (%) | |||
| Wildtype | 137 (96%) | 34 (83%) | 0.01 |
| Deleted | 6 (4%) | 7 (17%) | |
| Immune190, median (Q1, Q3) | 0.20 (0.15, 0.25) | 0.19 (0.15, 0.26) | 0.88 |
| Activated CD4, median (Q1, Q3) | −0.07 (−0.14, 0.01) | −0.02 (−0.07, 0.06) | < 0.001 |
| Activated CD8, median (Q1, Q3) | 0.15 (0.09, 0.24) | 0.19 (0.14, 0.29) | 0.02 |
| T regulatory cell score, median (Q1, Q3) | 0.07 (0.01, 0.13) | 0.05 (−0.02, 0.09) | 0.08 |
| Angiogenesis score, median (Q1, Q3) | 0.42 (0.35, 0.47) | 0.42 (0.35, 0.48) | 0.95 |
| Tertiary lymphoid structure, median (Q1, Q3) | 0.12 (0.08, 0.18) | 0.10 (0.04, 0.16) | 0.25 |
| PD-L2 expression, median (Q1, Q3) | 0.14 (−0.01, 0.32) | 0.07 (−0.11, 0.28) | 0.20 |
| Myeloid-derived suppressor cell expression, median (Q1, Q3) | −0.08 (−0.16, 0.01) | −0.10 (−0.22, −0.03) | 0.15 |
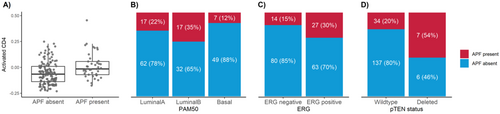
We next sought to validate prior studies that have shown Decipher can predict adverse pathologic features independent of clinical features, such as PSA levels. Scaled logistic regression after adjustment for logPSA showed that for each standard deviation increase in Decipher score, the odds of APF increased by 61% (OR 1.61, 95% CI 1.11–2.32, p = 0.01) (Figure 2).
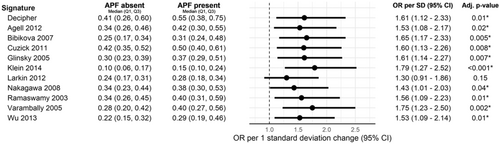
Finally, we took the genomic makers signatures that were associated with APF on comparative statistics and sought to establish an association with APF in the context of clinical features (PSA) on multivariate analysis. Scaled logistic regression after adjustment for logPSA showed APF was associated with PTEN gene expression loss, activated CD4 score, and ERG positive rate (Supplemental Table S2). Scaled logistic regression after adjustment for logPSA showed that multiple prognostic gene expression signatures are also associated with an increased risk of APF. In the scaled models, APF was associated with the gene expression signatures described by Agell, Bibikova, Cuzick, Glinsky, Klein, Nakagawa, Ramaswamy, Varambally, and Wu (Figure 2).
4 Discussion
Accurate risk stratification remains an integral part of prostate cancer management, particularly for patients considering active surveillance versus treatment. While traditional biomarkers and pathologic findings such as PSA and GG remain the cornerstone of risk stratification, there has been growing interest in incorporating genomic biomarkers into management decisions. While Decipher has previously been applied to low and favorable intermediate risk patients to predict APF, this association has not been established with other genomic scores. Therefore, we sought to compare association of various genomic signatures with APF in the context of clinical and pathologic variables.
In our relatively homogeneous cohort of primarily favorable intermediate risk men selecting surgical management, higher PSA, but not PSA density, tumor volume, or NCCN risk groups, was the only clinical factor significantly associated with APF at RP. In contrast, we found that multiple genomic signatures and classifiers were associated with APF. While the Decipher prostate genomic classifier has been previously calibrated and has established clinical cutoffs, many of these other signatures have not [8]. Therefore, we next sought to compare association of other genomic signatures with APF in the context of PSA levels. We performed scaled logistic regression adjusting for logPSA. We found that APF was associated with other previously published prognostic signatures, many that that were not originally designed for use in this population and most have not been studied previously in this context [13, 24]. This overall agreement across multiple genomic classifiers perhaps provides increased confidence when counseling patients on disease risk.
While genomic testing and molecular classification are commonly used in advanced prostate cancer, it has not been widely adopted yet in patients with lower risk disease. The relative incidence of adverse molecular features and potentially actionable genomic changes (e.g., AR-A, HR deficiency, PTEN loss, activated CD4) among patients with favorable disease is unknown. PTEN is one of the most common alterations in prostate cancer, occurring in 20–50% of primary tumors, and has been associated with adverse pathology at RP, shorter BCR free survival, and increased frequency of metastatic disease [34]. Furthermore, PTEN loss was predictive of grade reclassification in an AS cohort of low risk men. In our cohort, which is predominantly favorable intermediate risk disease, we find that PTEN loss is also predictive of APF at RP, and this relationship held when adjusting for logPSA. Increased CD4 activity has also been associated with adverse oncologic outcomes in prior studies, and we find similar results in this AS population [35]. This suggests that the immune system and tumor microenvironment may impact cancer development and progression. Finally, the previously referenced transcriptomic panels contain gene signatures that encompass a myriad of functions, including cell proliferation, cell differentiation, cell adhesion, signal transduction, immune modulation, and metabolism [24-29].
Our data suggests further research into the potential leveraging of these molecular signatures as therapeutic targets to reduce progression of prostate cancer for men on AS. For example, previous work has suggested that a short course of an advanced androgen receptor inhibitor could treat localized, favorable risk prostate cancer [36, 37]. The ENACT study demonstrated significant treatment effect in patients undergoing AS who received enzalutamide monotherapy, albeit with considerable side effects [38]. Genomic analysis has the potential to identify patients who may most benefit from these therapies and spare others from toxicity [39].
Limitations to this study include the combination of both biopsy and RP specimens for analysis. However, the concordance between Decipher biopsy and RP scores has been previously shown to be high [40]. In our data, consistent effect sizes were observed when modeling the odds of observing adverse pathology when limited to biopsy specimens and RP specimens (Supplemental Tables S3 and S4). There may be selection bias with inclusion of Decipher RP patients in enriching for those with adverse pathology. Furthermore, there is heterogeneity within our biopsy cohort, which is notable because there is evidence to suggest that genomic prediction from samples obtained from diagnostic versus surveillance biopsies are different [41]. Due to the low incidence of biochemical recurrence, metastasis, and prostate cancer-specific mortality in our cohort, as would be expected from an AS-eligible population, we were not able to perform meaningful analysis on these oncologic endpoints. However, APF does have implications in clinical decisions and is frequently used as a surrogate endpoint in this population of men with favorable disease. Furthermore, due to the low incidence of radical prostatectomy in the active surveillance eligible population, our sample size is relatively small, limiting statistical power of our study.
We report enrichment of adverse molecular features in AS-eligible patients who underwent RP and were found to have APF. This data can help providers risk stratify by providing both clinical and genomic variables associated with APF, as well as identify molecular-based systemic therapies that may benefit AS-eligible patients. Genomic biomarkers represent an exciting frontier and its incorporation in clinical trial design will elucidate the role of transcriptomics to move prostate cancer medicine further toward precision medicine.
Author Contributions
Jamie Michael, Eric V. Li, Mitch Huang, and Nicole Handa assisted in manuscript drafting and revisions. Nalin Kundu and Austin Ho assisted in data abstraction. Hiten D. Patel, Ridwan Alam, Edward M. Schaeffer, and Ashley E. Ross assisted in study planning and execution, as well as manuscript revisions. James A. Proudfoot, Sai Kaushik Shankar Ramesh Kumar, and Elai Davicioni assisted in statistical analysis.
Acknowledgments
The authors have nothing to report.
Conflicts of Interest
J.A.P. and E.D. are employed by Veracyte. A.E.R. is a consultant for Veracyte.
Open Research
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.