Prostate tumor RON receptor signaling mediates macrophage recruitment to drive androgen deprivation therapy resistance through Gas6-mediated Axl and RON signaling
Abstract
Background
Androgen deprivation therapy (ADT), or chemical castration, is the first-line therapy for prostate cancer; however, resistance leaves few treatment options. Prostatic tumor-associated macrophages (TAMs) have been shown to promote prostate cancer growth and are abundant in castration-resistant prostate cancer (CRPC), suggesting a role in promoting CRPC. We recently showed a tumor cell-intrinsic mechanism by which RON promotes CRPC. Given previous reports that RON alters prostate cancer cell chemokine production and RON-overexpressing tumors alter macrophage function, we hypothesized that a macrophage-dependent mechanism regulated by tumor cell intrinsic RON also promotes CRPC.
Methods
Using RON-modulated genetically engineered mouse models (GEMMs) and GEMM-derived cell lines and co-cultures with bone marrow-derived macrophages, we show functional and molecular characteristics of signaling pathways in supporting CRPC. Further, we used an unbiased phosphokinase array to identify pathway interactions regulated by RON. Finally, using human prostate cancer cell lines and prostate cancer patient data sets, we show the relevance of our findings to human prostate cancer.
Results
Studies herein show that macrophages recruited into the prostate tumor microenvironment (TME) serve as a source for Gas6 secretion which serves to further enhance RON and Axl receptor activation in prostate tumor cells thereby driving CRPC. Further, we show targeting RON and macrophages in a murine model promotes CRPC sensitization to ADT.
Conclusions
We discovered a novel role for the RON receptor in prostate cancer cells in promoting CRPC through the recruitment of macrophages into the prostate TME. Macrophage-targeting agents in combination with RON/Axl inhibition are likely to provide clinical benefits for patients with CRPC.
1 INTRODUCTION
Prostate cancer claimed an estimated 375,000 men worldwide in 2020.1 Since 1941, the first-line therapy for prostate cancer has remained androgen deprivation therapy (ADT; also known as chemical castration) which is sufficient for many patients unless resistance occurs.2 Because of resistance and resulting metastatic progression, prostate cancer continues to be a significant public health issue necessitating novel treatment strategies. For several cancers, immunotherapy has proved to be a successful treatment modality reigning in a new era of cancer immunotherapy. Altering the function of immune cell types in the microenvironment is thus a bona fide cancer treatment modality, and expansion of immune cell targeting treatments has the potential to improve prostate cancer patient outcomes.
Tumoral macrophage infiltration is implicated in more severe prostate cancer disease outcomes as it correlates with progression3 and significantly shorter disease-free survival following radical prostatectomy.4, 5 Further, preclinical models have detailed several roles for macrophages in prostate cancer via various mechanisms (e.g., inhibition of T-cell antitumor responses through protein nitration6) that support prostate tumor growth and metastatic potential.7, 8 Thus, macrophages within the tumor microenvironment (TME), also known as tumor-associated macrophages (TAMs), have demonstrable functions in supporting cancer progression. Macrophages exhibit a spectrum of functional plasticity with classically activated, proinflammatory, antitumor M1 macrophages on one extreme and alternatively activated, anti-inflammatory, protumor M2 macrophages on the other extreme.9
A novel driver of prostate cancer, of which our laboratory recently showed supports M2 macrophage activation in the prostate TME,10 is the RON receptor tyrosine kinase. However, mechanisms by which RON expression in prostate cancer TME utilizes TAMs remain unexplored. Recent work has detailed a tumor cell intrinsic function for RON in driving resistance to ADT.11-15 Given that RON has been reported to be overexpressed in the majority of castration-resistant prostate cancer (CRPC) patients, we report herein a newly identified mechanism for RON to promote CRPC through recruitment of macrophages in prostate tumors that supply Gas6, of which we are the first to report ligand activation of RON via Gas6. RON overexpressing prostate cancer cells both recruit macrophages and are more effective at utilizing macrophages to promote CRPC through sustained activation of RON and Axl. We also provide preclinical evidence that combining RON inhibition with macrophage depletion sensitizes castration-resistant tumors to castration therapy, providing a novel treatment option for men with CRPC.
2 MATERIALS AND METHODS
2.1 Immunohistochemistry and scoring
Murine subcutaneous tumor tissues were fixed in 10% formalin, paraffin-embedded, and cut into 5 μm sections before staining for BrdU (52925S; Cell Signaling), Ki67 (MA5-14520; ThermoFisher Scientific), TUNEL (Millipore In Situ Apoptosis Detection Kit), F4/80 (14 4801 85; eBiosciences), Arginase (610708; Becton Dickinson), iNOS (610328; Becton Dickinson) was performed as previously described.10 Hematoxylin and eosin (H&E) staining was performed as previously described.16 For BrdU analysis, mice were injected intraperitoneally with 200 µl 3 mg/ml BrdU in 0.9% normal saline 2 h before euthanasia. Samples with immunoglobulin G control antibody were used as negative controls. Images were taken of at least three fields per slide and all cells in the field were counted. Cells staining positively were divided by the total number of cells and then multiplied by 100 to obtain % positive cells/field. The average of at least three fields per slide was used as the % positive cells/field for that sample; at least three samples were used per genotype.
2.2 Western blot analyses
Cells were homogenized in RIPA buffer as described.17 Nuclear and cytoplasmic extracts were isolated by centrifugation and hypotonic lysis as described.18 Antibodies for analyses included: RON (SC-322), Androgen Receptor, (SC-815), Axl (SC-1096), Gas6 (SC-1935), and Tubulin (SC-5286) from Santa Cruz Biotechnology; Src (2110 S), phosphor-Src y416 (2101 S), Akt (4691 S), phosphor-Akt s473 (4060S), phospho-Axl y702 (5724S), and LAMIN A/C (4777S) from Cell Signaling Technologies; phospho-RON y1238/y1239 (AF1947, R&D); ACTIN (Cincinnati Children's Hospital Medical Center, Clone C4). Peroxidase-conjugated secondary antibodies (Jackson Laboratories) were applied, and membranes were developed using Pierce ECL2 Western Blotting substrate (ThermoFisher Scientific). Membranes were stripped using Restore Western Blot Stripping Buffer (ThermoFisher Scientific) before re-probing. The Proteome Profiler Mouse Phospho-RTK Array Kit (ARY014, R&D) was used on whole tumor lysates and performed according to the manufacturer's instructions.
2.3 Mouse models
Mice were maintained under specific pathogen-free conditions and experimental protocols approved by the University of Cincinnati IACUC. For prostate cell injections, 1.0 × 106 cells were injected subcutaneously into the flanks of 6−8-week-old male FVB mice as described.19, 20 Tumor growth was measured via calipers and volume was determined by the formula 0.5 × Length × Width2.21 Surgical castration was performed as described when tumors reached 1000 mm3.19, 20 For in vivo kinase inhibitor studies, mice were treated with 50 mg/kg/day BMS-777607 (Selleck Chemicals) or methylcellulose (vehicle) via oral gavage. Mice treated with clodrosome (Encapsula Nanosciences CLD8909) for macrophage depletion were injected with 200 µl intraperitoneally every 3 days. Hi-Myc mice (FVB-Tg(ARR2/Pbsn-MYC)7Key/Nci, Strain # 01XK8) were obtained through the mouse repository at the National Cancer Institute and crossed with ARR2Pb-RON strain B mice which were described previously.12, 22 Mice were euthanized and tissues were collected for analysis at 30 weeks of age.
2.4 Cell models
Myc-CaP19 and Pten-CaP223 murine cell lines were obtained from the laboratory of Charles Sawyers and the Memorial Sloan Kettering Cancer Center. For tumor formation, 1 × 106 Myc-CaP cells were injected subcutaneously into wild-type FVB male mice; once tumors reached 1000 mm3, the mouse was castrated. Myc-CaP-C cells were generated from a castration-resistant tumor formed following injection of Myc-CaP cell.11 All Myc-CaP cells were maintained in Dulbecco's modified eagle medium (DMEM) with 10% Cosmic Calf Serum and 1% gentamycin.19 The human prostate cancer cell lines LNCaP and C4-2B were obtained from ATCC and were maintained in RPMI-1640 with 10% fetal bovine serum (FBS) and 1% gentamycin. All cells were maintained at 37°C and 5.0% CO2. Bone marrow-derived macrophages (BMDMs) were isolated as previously described and cultured in DMEM supplemented with 10% FBS, 1% glutamine, 1% gentamycin, and 20 ng/ml M-CSF.24
2.5 Cell transfections
Stable polyclonal cell lines were generated by performing transfection with Lipofectamine 2000 (ThermoFisher Scientific) and selection in puromycin (Invitrogen, 5 µg/ml) or G418 (Invitrogen, 500 µg/ml). LNCaP Ctrl and LNCaP RON OE cells overexpressing human RON were generated as described.13 The RON gene was deleted in Myc-CaP cells using CRISPR/Cas9 technology as described previously to generate Myc-CaP RON KO1 cells.11 Short hairpin RNA (shRNA) constructs were purchased from Cincinnati Children's Hospital Medical Center (CCHMC) for knockdown of Axl (TRCN00000023311 for shAxl 1, TRCN00000023313for shAxl 2) or CCL2 (TRCN00000034470 for shCCL2-1, TRCN00000034473 for shCCL2-2) and knockdown was performed as previously described11 and nontargeting (NT) hairpins of scrambled sequences were employed as a control. Pten-CaP2 cells were knockdown for RON (as detailed in Brown et al.11) and were used as an additional model to interrogate the relationship between tumor cell RON expression and CCL2 production.
2.6 Viral transduction
Lentivirus short hairpin RNA (Open Biosystems) was used to target murine shRON (RMM3981-9590952), and nonsense shNT (RHS1764). The pCDH backbone and pCDH-CMV-EF1-PURO-RON full-length mouse RON cDNA expression vectors were utilized for control and RON overexpression. Transduction was performed as described.13, 18
2.7 In vitro cell growth assays and treatments
Three-dimensional growth assays were performed as described previously.11, 14 Briefly, 2 × 104 cells were plated on top of 1.0% agarose in six-well plates in media supplemented with cosmic calf serum (Complete; Thermofisher Scientific) or charcoal-stripped serum (CSS, Midsci). Cells were left untreated, treated with PBS (vehicle), Gas6 (R&D, 1 nM daily), or macrophage conditioned media (CM) (R&D, 1:10 every other day) daily. After 10 days, images of spheres were taken using a Zeiss Axiovert S100TV inverted microscope (Carl Zeiss Microscopy), and spheres >25 µm in diameter were counted using ImageJ software. The 25 μm threshold was established based on the average sphere size obtained for the control cells.
2.8 CM and cocultures
Macrophage CM was collected from 2 × 104 BMDMs seeded in a six-well plate. Twenty-four hours before collection, BMDM media was removed and serum-free media with 1% gentamycin was added. Media was aspirated, centrifuged at 200g for 10 min, and then passed through a 40 µM filter before being placed on cells at a ratio of 1:10. Gas6 depletion from BMDM CM was performed by rotating BMDM CM with 1 µg/ml Gas6 antibody (SC-1935) for 2 h at 4°C then by adding 30 µl/ml protein A/G agarose beads (SC-2003) and rotating for 2 h at 4°C. Following incubations with antibody and beads, the mix was centrifuged at 1000g for 5 min and the supernatant removed and used for treatment. Incubation with beads only served as the control. For sphere forming coculture assays, 2 × 104 epithelial cells and 2 × 104 BMDM cells were placed in a single well of a six-well plate.
2.9 Quantitative real-time (qRT)-PCR
RNA was extracted with TRIzol (Invitrogen), and cDNA was prepared using the High-Capacity cDNA Reverse Transcriptase kit (Applied Biosystems). Quantitative PCR was performed with 2X SYBR Green Master Mix (Roche Diagnostics) on a Mastercycler ep realplex4 (Eppendorf). Data were normalized to an 18S reference gene and analyzed by ΔΔCT. Primer sequences included: Murine Tmprss2 (Forward: 5'-AAGTCCTCAGGAGCACTGTGCA-3'; Reverse: 5'-CAGAACCTCCAAAGCAAGACAGC-3'), Murine Klkb1 (Forward: 5'-AAAGTCAGCGGACAACCTGGTG-3'; Reverse 5'-AGATGGTGCGACACACAAAGGC-3'), 18S (Forward: 5'-AGTCCCTGCCCTTTGTACACA-3'; Reverse: 5'-GATCCGAGGGCCTCACTAAAC-3'), Murine Gas6 (Forward: 5'-AGAACTTGCCAGGCTCCTACTCTTG-3'; Reverse: 5'-TCGCCCATCACAGTGGCAGGTATAG-3'), Murine Vegf (Forward: 5'-GCAGAAGTCCCATGAAGTGA-3'; Reverse: 5'-TCCAGGGCTTCATCGTTA-3'), Murine Ccl2 (Forward: 5'-TTAAAAACCTGGATCGGAACCAA-3'; Reverse: 5'-GCATTAGCTTCAGATTTACGGGT-3'), Murine Arginase (Forward: 5'-AGCATGAGCTCCAAGCC-3'; Reverse: 5'-CAGACCAGCTTTCCTCAGTG-3'), Murine iNos (Forward: 5'-GTTCTCAGCCCAACAATACAAGA-3'; Reverse: 5'-GTGGACGGGTCGATGTCAC-3'), Human PSA (Forward: 5'-TGCCCACTGCATCAGGAA-3'; Reverse: 5'-GCTGACCTGAAATACCTGGCC-3'), Human VEGF (Forward: 5'-GACAAGAAAATCCCTGTGGGC-3'; Reverse: 5'-AACGCGAGTCTGTGTTTTTGC-3'), Human TMPRSS2 (Forward: 5'-CCTCTAACTGGTGTGATGGCGT-3'; Reverse: 5'-TGCCAGGACTTCCTCTGAGATG-3').
2.10 Microscale thermophoresis
RON protein was tagged with GFP as described. After labeling, the protein was collected in lysate from 293T cells overexpressing the RON-GFP fusion protein using RIPA buffer. Nonfluorescent Gas6 (R&D, 885-GSB-050) or HGFL (R&D, 4306-MS) was titrated in a 1:1 dilution series (concentrations between 0.05 and 27.00 nM for Gas6, 0.004 nM and 4.00 nM for HGFL). Samples were loaded into Monolith™ NT.115 MST Standard Capillaries (NanoTemper Technologies) and measured using a Monolith NT.115 and MO.Control software at room temperature (LED/excitation power setting 50%, MST power setting 60%). Data were analyzed using MO.Affinity Analysis software (NanoTemper Technologies). Data were normalized using fraction bound binding.25
2.11 Migration assays
2 × 104 BMDMs were plated on top of a 24-well plate Transwell Permeable Support with an 8 µM pore size (Costar, 3422). 1 × 105 prostate cancer cells were placed in the bottom of the 24-well plate. BMDMs were allowed to migrate for 20 h, fixed in methanol for 20 min, then stained with 0.1% Crystal violet. Images were taken of the transwell insert and the total number of cells migrated was counted using Image J software.
2.12 Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM). Statistical significance was determined by performing Student's t test for pairwise comparisons or ANOVA for comparison of multiple groups using GraphPad Prism software (GraphPad Software). All in vitro experiments represent the average of at least triplicate experiments. Significance was set at *p < 0.05.
3 RESULTS
3.1 RON overexpression enhances Myc-driven prostate tumorigenesis
To examine the effects of epithelial RON overexpression in a genetic model of prostate cancer, we crossed mice that overexpress RON in the prostate epithelium (ARR2Pb-Ron) with the well-established Hi-Myc model of prostate cancer, termed Hi-Myc Pb-Ron. Overexpression of RON significantly increased prostate tumor weight at 30 weeks of age (Figure 1A) and resulted in an increased incidence of prostate adenocarcinoma (Figure 1B). Upon further characterization, Hi-Myc Pb-RON mice had prostates that exhibited increased proliferation as marked by BrdU staining and decreased cell death as shown by TUNEL staining (Figure 1C) with assessments performed on similar tumor-bearing areas within the respective prostates.
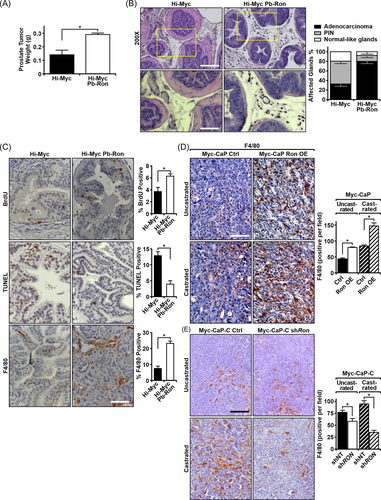
3.2 RON overexpression enhances F4/80+ cell recruitment into tumors in murine models of prostate cancer
Interestingly, we also observed changes in the TME between genotypes. While F4/80 staining of Hi-Myc prostate in our study is consistent with published studies that have examined macrophage recruitment in the Hi-Myc model,26, 27 in comparing similar tumor-bearing areas from Hi-Myc Pb-RON and Hi-Myc prostates, Hi-Myc Pb-RON prostates displayed an increased presence of F4/80+ cells (Figure 1C). Given the abundance of cells, we surmise the majority of these F4/80+ cells to be macrophages, as macrophages are reported to be the most abundant leukocyte within tumors.28 To determine whether this change in the TME was broadly applicable to other RON overexpressing prostate tumors, we assessed two previously characterized murine subcutaneous prostate cancer models.11 In these models, RON overexpression in either Myc-CaP cells (Myc-CaP RON OE) or in LNCaP cells was able to confer castration-resistant growth to control cells.11 In Figure 1D, we show that RON overexpression in Myc-CaP cells (Myc-CaP RON OE) also resulted in an increased presence of F4/80+ cells in subcutaneous tumors. IHC analyses were performed on Myc-CaP prostate tumors collected at similar sizes (Supporting Information: Figure S1A). Conversely, we previously showed that knockdown of RON in castration-resistant Myc-CaP-C cells resulted in both a delay in tumor growth following transplantation into pre-castrated mice as well as tumor regression following castration when implanted into intact FVB mice and castrated at 1000 mm3.11 As shown in Figure 1E, knockdown of RON in Myc-CaP-C cells decreased the presence of F4/80+ myeloid cells when compared to control Myc-CaP-C cells from prostate tumors collected at comparable sizes (Supporting Information: Figure S1B). Additional immunohistochemical and qPCR analysis revealed that Hi-Myc Pb-RON prostates have elevated expression of the M2 macrophage marker Arginase-1 relative to Hi-Myc control prostates (Supporting Information: Figure S1C−S1E).
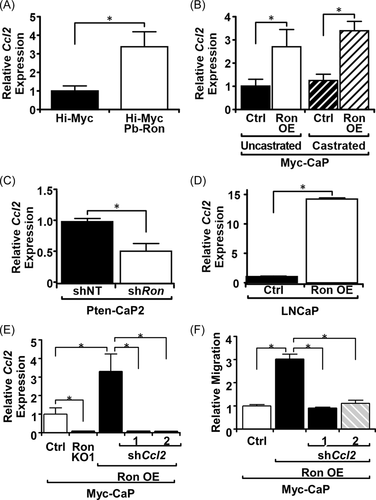
3.3 RON overexpression enhances CCL2 production, which is required to promote macrophage migration
RON overexpression has previously been established to regulate the synthesis and secretion of several cytokines.13 One of which, CCL2/MCP-1, has been established as a key regulator of macrophage chemotaxis. CCL2 mRNA expression was observed to be elevated in Hi-Myc Pb-RON prostates relative to Hi-Myc prostates (Figure 2A) and in Myc-CaP RON OE tumors relative to Myc-CaP Ctrl tumors (Figure 2B). Similarly, when RON was silenced in Pten-CaP2 cells (Figure 2C) or knocked out (KO1) in Myc-CaP cells (Figure 2E), CCL2 expression was decreased, while overexpressing RON in LNCap cells increased CCL2 expression (Figure 2D). To assess if the production of CCL2 has a function in influencing macrophage infiltration into RON overexpressing tumors, CCL2 was knocked down in Myc-CaP RON OE cells (Figure 2E), and cells were used for an in vitro migration assay measuring the migration of BMDMs toward prostate cancer cells. Knockdown of CCL2 reduced macrophage migration toward Myc-CaP RON OE cells to levels comparable to Myc-CaP Ctrl cells (Figure 2F), further supporting CCL2 as an effector molecule by which RON overexpressing prostate cancer cells recruit macrophages.
3.4 RON is required for BMDM-augmentation of tumorsphere growth
Macrophages have been established to play a critical role in the growth of prostate cancer and in the development of castration resistance.6-8 Further, RON overexpressing tumors, which exhibit increased macrophage infiltration, have previously been shown to be resistant to ADT.11 To assess whether RON overexpressing tumors utilize macrophages for growth in the absence of androgens, a three-dimensional-sphere culture assay was used to assess the growth of Myc-CaP cells with RON modulation with and without BMDMs.11 The addition of BMDMs increased sphere formation for both Myc-CaP control (Ctrl) and Myc-CaP RON OE cells; however, there was not a significant increase in sphere formation for Myc-CaP cells where RON was genetically deleted using CRISPR/Cas9 technology; images of spheres and results of quantitation are seen in Figure 3A. Additionally, the fold change for growth with and without the addition of macrophages was highest for Myc-CaP RON OE cells (2.42 for RON OE, 1.7 for Ctrl), suggesting that RON expression impacts the ability of prostate cancer cells to utilize macrophages for growth promotion.
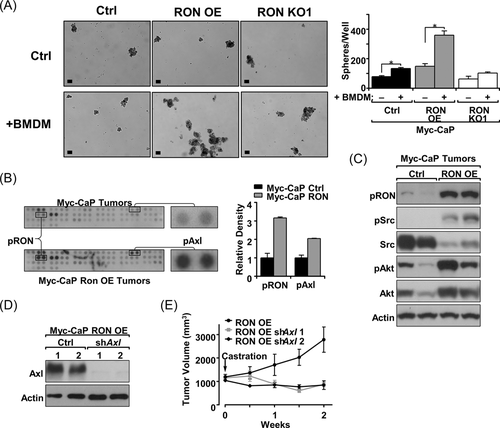
3.5 RON overexpressing prostate cancer cells require Axl to promote growth following castration
An unbiased phosphokinase array was used to determine what major signaling changes occur following RON overexpression in prostate tumors which may be a result of changes in the TME that allow growth under androgen deprivation (Table 1). The strongest phosphorylation induction in response to RON overexpression was for the Axl receptor (Figure 3B). Axl has been established as a driver of prostate cancer and in oral cancer was shown to be activated through myeloid cells of the TME.29 RON overexpressing Myc-CaP tumors were confirmed to have increased Axl signaling relative to control tumors as shown by increased Src and Akt phosphorylation (Figure 3C). To determine if Axl plays a functional role in RON overexpressing tumors for promoting castration resistance, Axl was knocked down in Myc-CaP RON OE cells and growth in vivo following castration was observed (Figure 3D,E). Tumors grown from Myc-CaP RON OE cell implantation with Axl knockdown (shAxl 1, shAxl 2) were observed to be sensitive to castration therapy, unlike their Myc-CaP RON OE ctrl counterparts continuing to grow upon castration (Figure 3E). These data indicate that Axl is a key player for RON overexpressing tumors to grow in the context of castration via this apparent tumor cell extrinsic mechanism. It is worth noting that while RON overexpression in Myc-CaP cells promotes growth under uncastrated and castrated conditions,11 Myc-CaP RON OE cells with an Axl knockdown grow similar to controls before castration but become sensitive to castration therapy (Figure 3E and Supporting Information: Figure S2). These studies suggest that Axl is important primarily for castration-resistant growth which may be due to the increased number of F4/80+ cells observed in the prostate tumors under castrated conditions with RON overexpression (Figure 1D) wherein higher levels of Gas6 would be available to activate Axl.
| Coordinatesa | Receptor | Fold change | Coordinatesa | Receptor | Fold change |
|---|---|---|---|---|---|
| A1, A2 | EGFR | 1.073708023 | B17, B18 | Tie-1 | 1.390132 |
| A3, A4 | ErbB2 | 0.839397091 | B19, B20 | Tie-2 | 1.068679 |
| A5, A6 | ErbB3 | 1.133019869 | B21, B22 | TrkA | 1.4348 |
| A7, A8 | ErbB4 | 1.759371593 | B23, B24 | TrkB | 1.882351 |
| A9, A10 | FGF R2 | 1.08292947 | C1, C2 | TrkC | 0.802591 |
| A11, A12 | FGF R3 | 0.793624668 | C3, C4 | VEGF R1 | 0.721265 |
| A13, A14 | FGF R4 | 1.264547344 | C5, C6 | VEGF R2 | 0.818611 |
| A15, A16 | Insulin R | 0.978055335 | C7, C8 | VEGF R3 | 0.851168 |
| A17, A18 | IGF-1R | 1.013540153 | C9, C10 | MuSK | 0.679367 |
| A19, A20 | Axl | 2.044932296 | C11, C12 | EphA1 | 1.137668 |
| A21, A22 | Dtk | 0.75016072 | C13, C14 | EphA2 | 1.152849 |
| A23, A24 | Mer | 0.50532455 | C15, C16 | EphA3 | 0.533973 |
| B1, B2 | c-Met | 1.609679 | C17, C18 | EphA6 | 0.896544 |
| B3, B4 | Ron | 3.16731 | C19, C20 | EphA7 | 1.125449 |
| B5, B6 | PDGF Ra | 1.231418 | C21, C22 | EphA8 | 0.924373 |
| B7, B8 | PDGF Rb | 1.57125 | C23, C24 | EphB1 | 0.961356 |
| B9, B10 | SCF R | 1.768606 | D1, D2 | EphB2 | 0.922031 |
| B11, B12 | Flt-3 | 1.498765 | D3, D4 | EphB4 | 1.611381 |
| B13, B14 | M-CSF R | 0.969688 | D5, D6 | EphB6 | 0.872447 |
| B15, B16 | c-Ret | 1.185835 |
- a Coordinates indicate the dot location for each receptor on the phosphoarray shown in Figure 3. Fold change normalizes the densitometric value from the phosphoarray performed using a Myc-CaP RON overexpressing tumor relative to the densitometric value from the phosphoarray performed using a Myc-CaP Ctrl tumor. A value greater than 1 indicates an increase of signal with RON overexpression, and a value less than 1 indicates a reduction in signal with RON overexpression.
3.6 Gas6 binds to RON and induces RON activation
Receptor tyrosine kinases can be activated through ligand binding or through receptor crosstalk with other receptors. In fact, RON has been shown to crosstalk with c-Met, PDGFR-β, IGF1-R, and others.30-32 To determine if RON overexpression induces Axl activation through receptor crosstalk, Myc-CaP and LNCaP cells were treated with the ligan for Axl (Gas6) or RON (HGFL) and phosphorylation of Axl and RON was measured. Interestingly, HGFL addition induced RON phosphorylation but did not induce Axl phosphorylation, indicating crosstalk is not a likely option (Figure 4A). However, the addition of Gas6 induced phosphorylation of both RON and Axl (Figure 4A). To determine if the phosphorylation of RON upon Gas6 stimulation requires Axl to be present, 293T cells, which have undetectable endogenous levels of Axl and RON, were treated with HGFL and Gas6. Following stimulation with either HGFL or Gas6 in control 293T cells, RON phosphorylation was not observed; however, stimulation with either HGFL or Gas6 in 293T cells where RON was exogenously expressed resulted in comparable levels of RON phosphorylation between the two ligands (Figure 4B).

3.7 Gas6 treatment induces nuclear localization of the androgen receptor (AR)
We previously reported that RON activation drives nuclear localization and activation of the androgen receptor under androgen deprivation, and stimulation of Myc-CaP cells with Gas6 under conditions of androgen deprivation resulted in increased AR nuclear localization (Figure 4C). Increased expression of the AR target genes, VEGF, TMPRSS2, and PSA, in both murine Myc-CaP and human LNCaP cells was also observed, with the greatest induction occurring in RON overexpressing cells (Figure 4D,E). These results indicate that the addition of Gas6 increases RON phosphorylation and activation; however, this does not appear to be through receptor crosstalk with Axl (Figure 4A−E). An alternative option is that Gas6 binds directly to RON to facilitate activation. To test this mechanism, microscale thermophoresis was used to measure the binding potential of both HGFL and Gas6 to a GFP-labeled RON receptor. As shown in Figure 4G, HGFL was shown to bind to RON with an EC50 of 0.0619 ± 0.0065 nM, which is similar to published data using sandwich ELISA analyses.33 Interestingly, Gas6 was also shown to bind to RON, with an EC50 of 0.5672 ± 0.0689 nM (Figure 4F). These results show the ability of Gas6 to bind to RON based on microscale thermophoresis studies. Finally, to test whether Gas6 binding to RON, rather than Axl, elicits expression of AR target genes, we treated Myc-CaP RON OE shAxl cells with Gas6 or HGFL (as a positive control) and found a similar induction of AR target genes (Tmprss2 and Klkb1) under Gas6 and HGFL stimulation conditions (Supporting Information: Figure S3). Taken together, these studies suggest that Gas6 binds to RON, even in the absence of Axl, and elicits AR nuclear translocation and transcription in prostate cancer cells.
3.8 Macrophage-produced Gas6 increases the growth of RON overexpressing prostate cancer cells under androgen-deprived conditions
After observing that receptor crosstalk was not likely the mechanism for Axl activation in the context of RON overexpression, we hypothesized that the increased presence of Gas6 in tumors may be responsible for inducing Axl activation. Our data show that prostate tumors from RON overexpressing cells have more F4/80+ cells (Figure 1). Moreover, RON overexpressing cells are more responsive to macrophages for the promotion of growth under castrate conditions (Figure 3). Further, others have shown that macrophages produce Gas6.34 Gas6 was observed to be upregulated at the mRNA level in Myc-CaP RON OE tumors relative to Myc-CaP Ctrl tumors (Figure 5A). Interestingly, there was no difference in the expression of Gas6 in Myc-CaP Ctrl and Myc-CaP RON OE cells, suggesting that Gas6 must be coming from another cell of the TME. To test if macrophage-derived Gas6 can promote the growth of RON overexpressing cells under androgen-deprived conditions, we added macrophage CM to various prostate cancer cells with and without Gas6 depletion under androgen-deprived conditions (Figure 5B,C). We observed that, like the coculture experiments in Figure 3A, the addition of BMDM CM promoted sphere formation of Myc-CaP Ctrl and RON OE cells, but no significant changes were observed in Myc-CaP RON KO1 cells (Figure 5C). Strikingly, Gas6 depletion reduced sphere formation in Myc-CaP RON OE cells to levels comparable to cell with no addition of BMDM CM (Figure 5C). Additionally, the exogenous addition of Gas6 to Myc-CaP RON OE cells exhibited a similar phenotype to the addition of BMDM CM, with no significant changes occurring in Myc-CaP Ctrl or Myc-CaP RON KO1 cells (Figure 5C). These data indicate Gas6 as a critical component secreted from macrophages for RON overexpressing cells to grow in the absence of androgens.
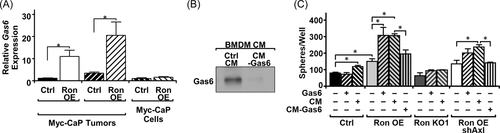
To determine the extent that macrophage secreted Gas6 supports prostate sphere formation of Myc-CaP RON OE cells through Axl activation, Myc-CaP RON OE shAxl cells were also treated with Gas6 and BMDM CM with/without Gas6 depletion. Interestingly, Gas6 addition and BMDM CM still promoted growth in Myc-CaP RON OE shAxl cells (1.7-fold), although not to the same extent as in Myc-CaP RON OE cells maintaining endogenous levels of Axl (2.1-fold) (Figure 5C). Additionally, Gas6 depletion from BMDM CM still resulted in a decrease in sphere formation to comparable levels to non-BMDM CM-treated cells (Figure 5C). These data demonstrate that Gas6 secreted from macrophages can activate both RON and Axl on prostate cancer cells for the promotion of growth under androgen-deprived conditions.
3.9 Macrophage depletion and RON inhibition sensitize castration-resistant tumors to castration therapy
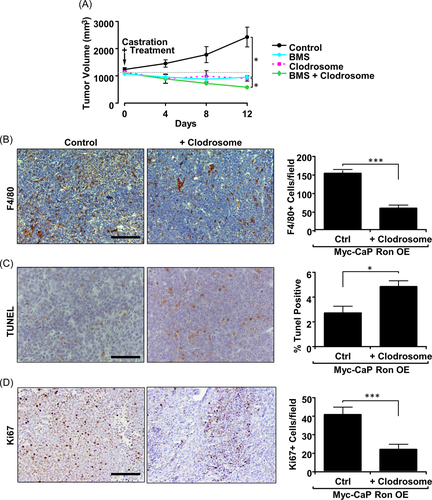
After establishing in vitro that RON overexpressing prostate cancer cells can utilize macrophages to promote growth under androgen-deprived conditions, we next sought to test the therapeutic implications of this relationship in vivo. This concept was tested through depletion of macrophages with clodrosome and/or inhibition of RON/Axl with BMS777607/ASLAN002 (BMS) in combination with castration therapy in mice with RON OE tumors.35 Myc-CaP RON OE tumor growth is dramatically reduced when macrophages are depleted in combination with castration therapy (Figure 6A). Treatment with clodrosome significantly reduced the number of F4/80+ cells in tumors as judged by F4/80 staining (Figure 6B). Moreover, clodrosome-treated mice exhibited increased tumor cell death and decreased cellular proliferation (Figure 6C,D). Interestingly, the reduction in tumor growth in the clodrosome-treated group was like the reduction in tumor growth in the BMS-treated group (Figure 6A). However, combined clodrosome and BMS treatment showed the largest response. The reasoning for this result is unclear; however, this may be because tumors with RON overexpression had been established and allowed to reach 1000 mm3 before castration, and thus RON-dependent alterations to macrophage recruitment/TME may have already been established. F4/80 staining of vehicle-treated tumors versus BMS-treated tumors (post-castration) showed no difference in F4/80+ staining, suggesting that CCL2 may be available at this time to allow for macrophage recruitment (Supporting Information: Figure S4A) or that the timing of BMS treatment may need to be altered to limit macrophage recruitment. In this context, the sustained presence of macrophages could have altered the function of other immune cells even if the GAS6/RON/Axl axis is ultimately disrupted, as we and others have shown.15 The combination of the two treatments may effectively reduce activation of both RON and Axl to sufficient levels for maximum tumor regression. RON and Axl inhibition in BMS-treated mice was also supported by IHC staining for phospho-RON (P-RON; Supporting Information: Figure S4B) and phospho-Axl (P-Axl; Supporting Information: Figure S4C). These data indicate that immunomodulatory agents targeting macrophages may have success in RON overexpressing castration-resistant tumors and efficacy may be enhanced when used in combination with RON/Axl inhibitors.
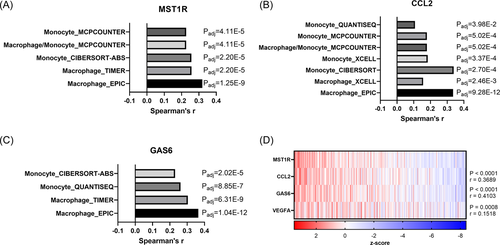
3.10 RON (MST1R) gene expression is correlated with macrophage/monocyte recruitment signatures and CCL2 and GAS6 gene expression in prostate cancer patient samples
Herein, we established that RON overexpression in prostate cancer cells promotes CCL2 production which can recruit macrophages to the TME. Macrophages in the TME secrete Gas6, further activating RON and Axl signaling. To examine this mechanism in the context of human prostate cancer, we analyzed patient-derived gene expression data sets. The Tumor Immune Estimation Resource (TIMER) tool was queried for MST1R, CCL2, and GAS6 revealing significant correlations between each gene and macrophage/monocyte recruitment supporting our model wherein RON-dependent CCL2 recruits Gas6-producing macrophages (Figure 7A−C). Next, we sought to determine the relationship between MST1R, CCL2, and GAS6 within human prostate cancers. A strong correlation is found within primary and metastasized human prostate tumors (Figure 7D−G). Importantly, this relationship is evident in neuroendocrine prostate cancer, a rare metastatic CRPC subtype (Figure 7G). These data further support our previous data demonstrating the role of RON in inducing CCL2 and ultimately Gas6 expression leading to macrophage infiltration and the progression of CRPC.
4 DISCUSSION
Currently, there is no curative therapy for men with CRPC, and immunotherapies that focus on altering T-cell activity have yet to produce significant results in CRPC patients, illustrating the need for novel approaches to treat men with CRPC. Previous studies have shown a pivotal role for tumor-infiltrating myeloid cells in the promotion of prostate cancer.15 Additionally, work from our laboratory and others have identified the RON receptor as a relevant driver of CRPC through tumor cell intrinsic activation of β-catenin and NF-kB that support androgen-independent activation of AR. Herein we demonstrate that RON overexpression in prostate tumor cells results in a restructuring of the TME with the increased presence of F4/80+ cells (Figure 1). Further, we show that the increased presence of F4/80+ cells in the TME is likely due to elevated synthesis and secretion of the macrophage chemoattractant protein CCL2 (Figure 2). Prior work from our group illustrated a novel role for RON in promoting endothelial cell recruitment to the prostate TME through the secretion of angiogenic chemokines; however, CCL2 was not implicated in this process.13 Other reports have shown that CCL2 production in prostate cancer cells induces infiltration of myeloid cells and promotes CRPC; however, CCL2 induction was driven by the stimulation of WNT5a through MAPK signaling.36 Given our previous work showing the requirement of β-catenin for restoration of AR activity under androgen deprivation, β-catenin may play a dual function of regulating CCL2 expression and AR activity. Together, our recent data and prior work demonstrate that RON overexpressing prostate tumors structure the microenvironment to have higher presence of endothelial cells and myeloid cells compared to low RON tumors, and this structure increases prostate tumor growth potential and primes the tumor for ADT resistance.
Our data also reveal a novel role for macrophages in prostate cancer in facilitating the activation of both RON and Axl signaling through the secretion of Gas6 (Figures 3 and 4). Axl has been shown to play a significant role in prostate cancer, as silencing Axl in prostate cancer cell lines resulted in the reduction of proliferation, migration, and invasion properties37; however, Axl signaling has not yet been directly linked to castration resistance or AR activation. Additionally, Gas6 stimulation has been shown to induce AR activation through activation of the nonreceptor tyrosine kinase Ack1.38 Interestingly, the cell surface receptor responsible for mediating the effects of Gas6 on AR through Ack1 was not detailed. Consistent with this report, we also observed that Gas6 stimulation induces activation of AR signaling, although we observed that this effect was enhanced with RON overexpression (Figure 4). Interestingly, Ack1 kinase is capable of activating NF-κB signaling.39 Although RON has not been shown to activate Ack1 kinase, Ack1 has been previously shown to be activated by other RTKs. It is therefore plausible that Gas6 stimulation results in a RON-Ack1-NF-κB-AR signaling cascade that is capable of driving growth in the absence of androgens. This mechanism has not yet been explored.
Through our analysis of the interplay between RON and Axl signaling, we made the novel discovery that Gas6 binds to and induces activation of RON in addition to human-associated data linking Gas6 and RON expression in CRPC (Figures 4 and 7). Multiple receptors have been identified that bind to Gas6 (Axl, Tyro3, Mer); however, the only known ligand for RON had been HGFL.40 This novel relationship may help explain several previously unknown functions of RON, which up until this point have simply been referred to as HGFL-independent functions. Previous studies in both breast and prostate cancer have utilized HGFL knockout mice to show that HGFL has specific roles in RON-driven tumorigenesis.14, 16 Interestingly, in breast cancer, cell spreading and survival have been determined to be HGFL-independent functions of RON.41 Further work will be needed to determine if any of these functions are Gas6-specific functions. It is interesting to note that when comparing the microscale thermophoresis (MST) data between the HGFL/RON and Gas6/RON interactions, it was determined that Gas6 has an almost 10-fold higher EC50 than HGFL. There are several potential reasons for this, each of which needs to be further evaluated. The HGFL used for MST contains a cysteine to alanine substitution at position 672, which increases the bioactivity of the recombinant protein. Although our measurements for the Gas6-RON ec50 are still threefold higher than published studies using a non-C672A substituted form of HGFL (0.5672 nm vs. 0.18 nm).33 Moreover, HGFL is secreted as an inactive precursor protein that needs to be cleaved to bind to RON and Gas6 does not require a cleavage event.42 When cell surface proteases are limited, binding of Gas6 to RON may be preferential to HGFL. Additionally, Tyro3 and Mer also bind to Gas6; however, their binding affinity is greatly improved in the presence of phosphatidyl serine.43 It is possible that the presence of another molecule may aid in Gas6 binding to RON. These potential avenues will need to be explored as the relationship between Gas6 and RON is further characterized.
Our data show that RON overexpression in prostate cancer alters and utilizes components of the TME to promote CRPC growth. We hypothesize that disruption of this microenvironment has the potential to improve patient outcomes for those suffering from CRPC. These preclinical data indicate that RON overexpressing prostate tumors are directly supported by macrophages for sustained growth under androgen deprivation (Figure 7). Macrophage-targeted approaches of immunotherapy for cancer have recently gained traction as the anti-CSF1R monoclonal antibody Axatilimab (SNDX-6352) is currently under investigation in two Phase I clinical trials for cholangiocarcinoma and metastatic solid tumors. Based on our results, we hypothesize that patients with RON overexpressing prostate cancer can be sensitized to ADT under a macrophage targeting treatment, such as via Axatilimab. Additionally, we hypothesize that disruption of RON/Axl signaling through the multi-kinase inhibitor BMS777607 used herein can also sensitize patients with CRPC to ADT. BMS777607 has recently completed a Phase I clinical trial for advanced solid tumors, and our preclinical work illustrates a need to pursue the use of drugs such as this in prostate cancer patient studies.44 Thus, BMS777607 and Axatilimab are exciting prospective agents for the treatment of CRPC.
5 CONCLUSIONS
Consistent with previous findings employing other genetically engineered mouse models of prostate cancer, RON overexpression promotes Myc-driven prostate cancer progression including enhanced tumoral recruitment of F4/80+ cells. RON overexpression in prostate cancer cells leads to enhanced CCL2 production, which is required for RON-dependent recruitment of macrophages. Macrophages sustain RON and Axl activation via Gas6 production, which drives prostate cancer growth under androgen-deprived conditions. A combination of RON/Axl inhibition and macrophage depletion sensitizes CRPC tumors to ADT.
ACKNOWLEDGMENTS
We would like to thank Glenn Doermann for his assistance in figure preparation and Madison Nashu, Jennifer Bourn, Camille Sullivan, and Andrew Paluch for technical assistance. This study was supported by the United States Department of Veterans Affairs research grant 1IOBX000803 (S. E. W.); National Institutes of Health Grants T32 CA117846 (S. E. W., A. J., N. E. B.), F31-CA200390 (N. E. B., S. E. W.), F31-CA228373 (B. G. H., S. E. W.), CA125379 (S. E. W.) and CA239697 (S. E. W.).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Samples from the publically available TGCA Pan Cancer data set were evaluated with the Tumor Immune Estimation Resource (TIMER) tool to reveal correlations between noted genes.