Aqueous metabolome of tissue-specific conditional Pten-knockout mouse prostate cancer and TRAMP neuroendocrine carcinoma
Sangyub Kim, Li Li, and Jinhui Zhang contributed equally to this study and they are co-first authors.
Preliminary results were communicated at the American Association for Cancer Research annual meeting in Washington, DC, April 2017 (Jinhui Zhang, Li Li, Sangyub Kim, et al., Integrative Analysis of Transcriptomic, Proteomic, and Metabolomic Data of Pten-Knockout Carcinogenic Mouse Prostate).
Abstract
Background
Metabolic reprograming is now a recognized hallmark of cancer. The prostate-specific phosphatase and tensin homolog deleted on chromosome 10 (Pten) gene-conditional knockout (KO) mouse carcinogenesis model is highly desirable for studying prostate cancer biology and prevention due to its close resemblance of primary molecular defects and histopathological features of human prostate cancer. We have recently published macromolecular profiling of this model by proteomics and transcriptomics, denoting a preeminence of inflammation and myeloid suppressive immune cell features. Here, we performed metabolomic analyses of Pten-KO prostate versus wild type (WT) counterpart for discernable changes in the aqueous metabolites and contrasted to those in the TRAMP neuroendocrine carcinoma (NECa).
Methods
Three matched pairs of tissue-specific conditional Pten-KO mouse prostate and WT prostate of litter/cage-mates at 20–22 weeks of age and three pairs of TRAMP NECa versus WT (28–31 weeks) were profiled for their global aqueous metabolite changes, using hydrophilic interaction liquid chromatography-tandem mass spectrometry.
Results
The Pten-KO prostate increased purine nucleotide pools, cystathionine, and both reduced and oxidized glutathione (GSH, GSSG), and gluconate/glucuronate species in addition to cholesteryl sulfate and polyamine precursor ornithine. On the contrary, Pten-KO prostate contained diminished pools of glycolytic intermediates and phosphorylcholine derivatives, select amino acids, and their metabolites. Bioinformatic integration revealed a significant shunting of glucose away from glycolysis-citrate cycle and glycerol-lipid genesis to pentose phosphate cycle for NADPH/GSH/GSSG redox and pentose moieties for purine and pyrimidine nucleotides, and glycosylation/glucuronidation. Implicit arginine catabolism to ornithine was consistent with immunosuppression in Pten-KO model. While also increased in cystathionine-GSH/GSSG, purine, and pyrimidine nucleotide pools and glucuronidation at the expense of glycolysis-citrate cycle, the TRAMP NECa increased abundance of many amino acids, methyl donor S-adenosyl-methionine, and intermediates for phospholipids without increasing cholesteryl sulfate or ornithine.
Conclusions
The aqueous metabolomic patterns in Pten-KO prostate and TRAMP NECa shared similarities in the greater pools of cystathionine, GSH/GSSG redox pair, and nucleotides and shunting away from glycolysis-citrate cycle in both models. Remarkable metabolic distinctions between them included metabolisms of many amino acids (protein synthesis; arginine-ornithine/immune suppression) and cholesteryl sulfate and methylation donor for epigenetic regulations.
1 INTRODUCTION
Activation of oncogenes and/or loss of tumor suppressor genes alter the metabolism of cancer cells to support their growth and survival.1 Cancer metabolic reprogramming is now recognized as a hallmark of cancer.2 The phosphatase and tensin homolog deleted on chromosome 10 gene (PTEN) loss is a frequent early pathogenetic change and a major oncogenic driver in human prostate cancer (PCa).3 The loss of PTEN alters its pivotal role in regulating various biological functions, including signal transductions, macromolecular alterations (engine), and metabolic reprograming (fuels).4-6
Metabolomic profiling allows an understanding of the global metabolic milieu changes associated with and perhaps even driving a particular cancer pathophysiology.7 The prostate-specific Pten-conditional knockout (KO) mouse carcinogenesis model is highly desirable for studies of prostate cancer biology and prevention due to its close resemblance of primary molecular defect and many histopathological features of human prostate cancer including androgen response, disease progression from prostatic intraepithelial neoplasia to adenocarcinoma, and a strong immunosuppressive tumor microenvironment (TME).8, 9 We have recently published macromolecular profiling of this model by proteomics and transcriptomics10 and contrasted with TRAMP and c-Myc transgenic mouse models of prostate carcinogenesis. Based on macromolecular omics as well as literature reports of Pten/AKT-mTOR signaling, we implicitly presumed the hypothesis that Pten-KO prostate undergoes altered global metabolism conducive for carcinogenesis and that some of the metabolic changes may be specific when compared to a different model of prostate carcinogenesis by different onco-driver such as the TRAMP model. Here we examined the aqueous metabolite metabolomics of Pten-KO mutant prostate to profile and incorporate metabolic disturbances into the Pten-KO driven carcinogenesis process. We applied the same metabolomics platform to the TRAMP model and compared and contrasted between these two pathogenetic models.
2 MATERIALS AND METHODS
2.1 Source of biospecimens
As described in detail previously,10 the mouse tissues were provided by the Mouse Model Repository for Cancer Biomarker Discovery led by Christopher J. Kemp, PhD, at the Fred Hutchinson Cancer Research Center, sponsored by National Cancer Institute Mouse Proteomics Initiative (http://proteomics.cancer.gov/programs/completed/mouseinitiative10). Tissues were collected and cryopreserved from mice of Pten-KO genotype and their wild type (WT) litter-/cage-mates in 129 background from April 2005 to April 2007.
The frozen tissue specimens were shipped to Texas Tech University Health Sciences Center School of Pharmacy, Amarillo, on dry ice (received on August 2, 2011) and stored at −80°C until analyses. The litter-/cage-mate paired tissues allowed comparison of the genotypic differences with strict control of housing and environmental factors, breeding stock, and age variations among the Pten-KO and WT mice. For the Pten-KO and matched prostate specimens obtained from mice aged 20–22 weeks, we divided the frozen tissues into smaller portions and used the leftover potions from the mRNA microarray omics for the metabolomics. The extraction protocols for each omic were unique to its platform and had to be performed separately. As described in our previous paper,10 the proteomics analyses were carried out on a different batch of WT and KO matched pairs at the younger age of 12–15 weeks. Due to the frozen nature of tissues obtained from the National Cancer Institute (NCI) repository, histology was not available. However, the pathology was assumed to involve invasive adenocarcinoma based on published work of the originator laboratory of Hong Wu, PhD,8 with significant infiltration of myeloid-derived immune cells and inflammatory cells.9
By the same procurement procedure as the Pten-KO pairs, three pairs of frozen TRAMP tumor and WT prostate in litter/cage-mates of 28, 29, and 31 weeks of age were also obtained. The matched pairs of mice were sacrificed on respective dates when a palpable pelvic mass was detected in the transgenic sibling. Similarly, lack of histology information, the TRAMP tumors were presumed poorly differentiated neuroendocrine carcinomas (NECa) based on literature from groups of authoritative experimental/veterinary pathologists 11 and our own.12 Due to the frozen nature of tissues as described above, we were not able to obtain specific lobes for the WT prostate of either model. We presume the prostate tissues sampled in our metabolomics work were mixture of different lobes.
2.2 Tissue sample extract preparation
The frozen tissue samples were homogenized in 30 volumes (v/w) of precooled (−80°C) 80% methanol in water and stored at −80°C for 16 h. The homogenates were centrifuged at 14,000 g for 10 min in a refrigerated centrifuge at 4°C. The supernatants were saved at −80°C and analyzed by the following protocol.
2.3 Metabolomics
The protocol published by Yuan et al.13 was adapted to profile aqueous endogenous metabolites from the tissue homogenate supernatants prepared in 80% methanol/water. The platform used hydrophilic interaction liquid chromatography with positive/negative ion switching to monitor 289 Q1/Q3 transitions from a single LC-MS/MS acquisition with a 5 ms dwell time and a 1.55 s cycle time. A UHPLC system (Nexera, Shimadzu) was coupled to a hybrid triple quadruple linear ion trap mass spectrometer (QTRAP 5500, AB Sciex). Chromatographic separation was performed on an XBridge BEH Amide Column (hydrophilic interaction liquid chromatography, 100 × 4.6 × 3.5 µm, Waters). The column oven temperature was kept at 40°C.
To ensure sensitive detection within the linear dynamic range, each extract was analyzed by multiple UHPLC-MS/MS runs with the injection volume of 10 and 1 µl. The abundance of each endogenous metabolite varied over a wide range, some of them were not able to be detected when 1 µl extract was injected; whereas the signal of some metabolites was saturated when 10 µl extract was injected. For 1 µl volume injections, all were done in duplicate. For 10 µl volume injections, most was done in duplicates, some in triplicates. The identification of metabolites was performed as per the Yuan report.13
2.4 Data curation and statistical metrics
The average/mean of the replicate measurements (peak area) was calculated and used for computation of the paired ratio (indicative of relative abundance) of each LC-MS/MS verified metabolite between Pten-KO or TRAMP prostate tumor and respective WT litter-/cage-mate. One-tailed, paired t test was administered on the peak areas in matched pairs (null hypothesis difference of tumor and WT pairs = 0) or on log-transformed peak area (null hypothesis ratio = 1 or logTumor-logWT = 0). Given the small number of pairs and inherent large variance among individual mice for metabolite analyses, p ≤ 0.1 or one-tail log paired p ≤ 0.1was used as statistical threshold for differential abundance for metabolites between tumor and WT litter-/cage-mate samples and for inclusion in enrichment analyses. For exception in the Pten-KO model, nine metabolites of clinical relevance and with the ratio changes for all three pairs concordant in direction were included for analyses.
2.5 Bioinformatic and metabolic enrichment analyses
We performed the statistical analysis and the enrichment analysis utilizing MetaboAnalyst 4.0 (link: www.metaboanalyst.ca/).14 The partial least-squares discriminant analysis (PLS-DA) model was used to highlight differences between Pten-KO or TRAMP tumors versus respective WT mouse prostate. For the creation of the heat maps, the selected metabolites were also hierarchically clustered by distance measure using Pearson's correlation and clustering algorithm using average linkage (clustering uses the centroids of the observations). The values were scaled metabolite-wise in columns for heatmap visualization. For the metabolic enrichment analyses, location-based metabolite sets and pathway-associated metabolite sets in MetaboAnalyst's enrichment analysis (MSEA) module were selected as a metabolite library for MSEA. The metabolic pathway map of the selected differential metabolites and significantly changed metabolic pathways was analyzed and visualized by iPath 3.0 software (https://pathways.embl.de/ipath3.cgi).
3 RESULTS
3.1 Aqueous metabolome of Pten-KO prostate
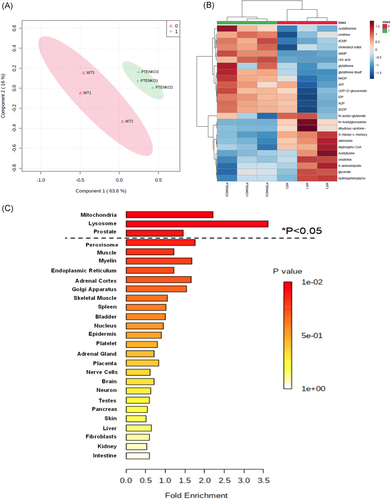
Using the UHPLC-MS/MS platform, we identified 177 endogenous metabolites in the mouse prostate aqueous extracts (Table S1, Raw data). With the exception of a detectable increase of cholesteryl sulfate, it was apparent for a lack of detection of hydrophobic lipids, fatty acids, cholesterol, and cholesteryl fatty acid esters. Table 1 summarized 44 selected metabolites (20 greater and 24 less in Pten-KO prostate than in WT prostate) that met criteria of statistical threshold or concordance of change directions described in Section 2, 2 (more details in Table S1, select metabolites). A supervised PLS-DA of the variance of these select metabolites between the two genotypes showed an obvious separation of WT prostate tissue (green crosses) and Pten-KO mutant prostate (red triangles) (Figure 1A). The heatmap of the top 25 metabolites (14 more abundant, 11 less abundant than WT) (Figure 1B) showed distinctive clustering of the metabolites between the genotypes.
| Metabolite | Pten-KO/WT ratio (n = 3) |
Pten-KO/WTa |
TRAMP/WT ratio (n = 3) |
Human PCa18 |
Human PCa17 |
||
|---|---|---|---|---|---|---|---|
| Increased in Pten-KO | Mean | SD | Mean | SD | |||
| ADP | 7.06 | 9.00 | 2.85 | 2.11 | 1.43 | ||
| ATP | 17.04 | 25.33 | 1.64 | 0.74 | |||
| IDP | 7.83 | 10.46 | 1.93 | 1.62 | 0.46 | ||
| dAMP | 1.95 | 0.88 | 4.68 | 1.71 | |||
| dCMP | 2.19 | 0.60 | Missing data | ||||
| dGDP | 7.58 | 9.95 | 1.59 | 0.64 | |||
| dGTP | 20.80 | 31.91 | 1.73 | 0.78 | |||
| Uric acid | 4.77 | 3.23 | 1.34 | 0.74 | 0.59 | 0.8 | 0.95 |
| Aminoimidazole carboxamide ribonucleotide | 1.29 | 0.20 | Missing data | ||||
| Cytosine | 1.53 | 0.25 | 0.05 | 0.01 | |||
| Cholesteryl sulfate | 4.51 | 1.64 | 1.19 | 0.35 | 0.21 | ||
| Cystathionine | 10.11 | 11.67 | 8.04 | 1.94 | |||
| Glutathione (GSH) | 1.55 | 0.61 | 0.68 | 1.72 | 0.41 | 1.14 | |
| Glutathione disulfide (GSSG) | 2.26 | 1.13 | 2.50 | 0.93 | 0.96 | ||
| NADP+ | 5.83 | 4.81 | 0.38 | 1.07 | 1.12 | 5.1 | |
| Ornithine | 3.75 | 2.47 | 2.50 | 1.76 | 1.42 | ||
| Pantothenate | 1.48 | 0.39 | 0.36 | 0.24 | |||
| p-Hydroxy-benzoate | 1.52 | 0.18 | 1.01 | 0.10 | |||
| d-gluconate | 1.50 | 0.18 | 1.12 | 0.36 | |||
| UDP-d-glucuronate | 5.03 | 5.39 | 2.22 | 0.70 | |||
| Decreased in Pten-KO | Mean | SD | Mean | SD | |||
| 4-Amino-butyrate | 0.22 | 0.16 | 1.95 | 1.78 | 1.00 | ||
| N-Acetyl-glucos-amine | 0.15 | 0.07 | 0.40 | 0.11 | 0.13 | 2.5 | |
| N-Acetyl-l-alanine | 0.44 | 0.24 | 2.41 | 0.58 | 0.28 | ||
| N-Acetyl-glutamate | 0.47 | 0.06 | Missing data | 0.4 | |||
| Adenosine | 0.39 | 0.28 | 0.28 | 0.16 | 0.15 | 0.6 | 0.91 |
| Acadesine | 0.32 | 0.23 | 1.63 | 0.35 | |||
| Guanine | 0.55 | 0.34 | 1.24 | 0.47 | |||
| Creatinine | 0.18 | 0.08 | 0.66 | 0.21 | 0.21 | 0.99 | |
| Creatine | 0.62 | 0.19 | 0.77 | 0.12 | |||
| Dephospho-CoA | 0.43 | 0.28 | Missing data | ||||
| Dihydroxy-acetone-phosphate | 0.26 | 0.11 | 0.33 | 0.13 | |||
| Glycerate | 0.22 | 0.10 | 1.48 | 0.23 | 0.02 | 1.04 | |
| Glycerophospho-choline | 0.58 | 0.32 | 0.37 | 0.09 | |||
| Phosphoryl-choline | 0.24 | 0.08 | 0.23 | 0.36 | 0.14 | ||
| Hydroxyphenyl-pyruvate | 0.32 | 0.13 | 1.60 | Missing data | |||
| Myo-Inositol | 0.66 | 0.32 | 0.39 | 0.16 | |||
| Nicotinamide- ribotide | 0.55 | 0.26 | 0.38 | 0.09 | |||
| Oxaloacetate | 0.86 | 0.06 | 0.61 | 0.04 | |||
| Sarcosine | 0.61 | 0.30 | 0.96 | 0.30 | |||
| sn-Glycerol-3-phosphate | 0.46 | 0.13 | 0.32 | 0.41 | 0.16 | 3 | 1.59 |
| S-Ribosyl-l-homocysteine | 0.27 | 0.16 | 0.09 | 0.11 | |||
| Trehalose-6-Phosphate | 0.47 | 0.45 | 1.41 | 0.15 | |||
| Trehalose | 0.50 | 0.06 | 1.02 | 0.11 | |||
- Note: Boldface metabolites and their fold ratio values met one or both p-value threshold.
- a Zabala-Letona.16 We selected average 6-month data to compare with our data.
Some metabolites were remarkably increased in the Pten-KO prostate tumors (Table 1). They included many ribosyl- and deoxyribosyl nucleotides (ADP, ATP, IDP, dAMP, dCMP, dGDP, dGTP), cytosine and nucleotide derivatives (AICA-ribonucleotide, NADP+, UDP-d-glucuronate), uric acid (a purine metabolite), glutathione (GSH) and its oxidized disulfide (GSSG) and key cysteine precursor cystathionine. Beside the increased abundance of cholesteryl sulfate, Pten-KO prostate contained a much-elevated level of ornithine, a known precursor to polyamines and a product of arginase to catalyze the release of urea from arginine (urea cycle).
On the other hand, metabolites with diminished abundance in the Pten-KO prostate (Table 1) included amino acid derivatives (N-acetyl-l-alanine, N-acetyl-glutamate, acetyl-lysine, S-ribosyl-l-homocysteine), purine base or nucleoside (adenosine, guanine), glycolysis intermediates, phosphorylcholine intermediates as well as other arginine and glycine metabolites such as creatine, creatinine, and sarcosine.
3.2 Bioinformatic analyses of Pten-KO prostate metabolome data set and integration with key proteomic and transcriptomic changes
We used MESA to identify possible pathways and biologically meaningful patterns enriched in the metabolomic data (Tables 2 and S2). The functional enrichment analysis yielded nine significant metabolic pathways (two upregulated and seven downregulated pathways) with p < 0.05 (Table 2). Purine metabolism and glutathione metabolism were the two upregulated pathways. The seven downregulated pathways were glycerophospholipid metabolism; arginine and proline metabolism; glycine, serine, and threonine metabolism; alanine, aspartate, and glutamate metabolism; glycerolipid metabolism, glycolysis, or gluconeogenesis; and pyruvate metabolism. Location-based metabolite sets MESA predicted that these metabolic pathways were mainly associated with mitochondria, lysosome, and prostate (Figure 1C). Not surprisingly, mitochondria and, to a less extent, lysosomes are the major subcellular organelles for providing building blocks as well as regulators of metabolic flux and redox activity in the cell.15
| Metabolism pathway | Direction of change | p value | Total | Hits | Selected metabolites altered in the pathway |
|---|---|---|---|---|---|
| Purine metabolism | Up | 3.02E–07 | 92 | 8 | AICA-riboside, ATP, ADP, dAMP, IDP, uric acid, dGDP, dGTP |
| Glutathione metabolism | Up | 2.18E–04 | 38 | 4 | Glutathione, oxidized glutathione, NADP, ornithine |
| Glycerophospholipid metabolism | Down | 6.94E–04 | 39 | 4 | Glycerol 3-phosphate, dihydroxyacetone phosphate, phosphorylcholine, glycerophosphocholine |
| Arginine and proline metabolism | Down | 0.00117 | 77 | 5 | N-Acetyl-l-alanine, creatine, creatinine, gamma-aminobutyric acid, sarcosine |
| Glycine, serine and threonine metabolism | Down | 0.00154 | 48 | 4 | Glycerate, dimethylglycine, sarcosine, creatine |
| Alanine, aspartate and glutamate metabolism | Down | 0.00198 | 24 | 3 | l-Alanine, gamma-aminobutyric acid, oxalacetic acid |
| Glycerolipid metabolism | Down | 0.00457 | 32 | 3 | Glycerol 3-phosphate, glycerate, dihydroxyacetone phosphate |
| Glycolysis or Gluconeogenesis | Down | 0.0435 | 31 | 2 | Oxalacetic acid, dihydroxyacetone phosphate |
| Pyruvate metabolism | Down | 0.0461 | 32 | 2 | Oxalacetic acid, dihydroxyacetone phosphate |
- Note: The bold face highlights a metabolite met statistical significance thereshold for different between genotypes. Plain face showed change concordant in the direction (i.e., either all increased or all decreased) for all three pairs. These “exception” were included due to their clinical relevance, as noted in text.
Based on mammalian biochemical reactions, Figure 2 mapped the majority of detected metabolite changes (bold for increased in Pten-KO; shaded white for decreased in Pten-KO) within the interconnected metabolic pathways, shunts, and cycles, focusing on glucose fates, GSH/GSSG, and their constituent amino acids glutamate, cysteine and glycine, and arginine/ornithine/urea cycle. The Pten-KO prostate shunted glucose away from glycolysis, as indicated by decreased dihydroxy-acetone-phosphate and glycerate and decreased glycerol/choline pool (decreased glycerol-3-phosphate, glycerophosphocholine, and phosphorylcholine), and from the flux through the citrate cycle, as indicated by diminished supply of acetyl-CoA to the mitochondria (decreased dephopsho-CoA, and decreased acetylation of glutamate and select amino acids and glucosamine) and a modest decrease of oxaloacetate. Instead, glucose was channeled toward pentose phosphate cycle (increased gluconate) for NADPH production and for the pentose moieties essential for anabolic synthesis or salvage of purine and pyrimidine nucleotides for nucleic acids crucial cell proliferation; and to the glucuronidation pathway for Phase II drug metabolism (increased UDP-glucuronate). The Pten-KO prostate increased the overall synthesis of GSH and redox fluxes of GSH/GSSG pair. Increased cystathionine would fuel a steady supply of cysteine for GSH synthesis. Another key amino acid glycine for GSH synthesis was likely made more available by diversion away from its S-adenosyl-methionine-driven methylation to generate sarcosine and the production of creatine via arginine:glycine aminotransferase (AGAT). For GSH/GSSG redox and GSH driven functions, we have reported increased protein abundance of glutathione disulfide reductase (GSR), glutathione S-transferases omega-1 (GSTO1) and A4 (GSTA4) and mRNA increases of Gsto1, glutathione peroxidase 2 gene (Gpx2), and hexose-6-phosphate dehydrogenase (H6pd) in the Pten-KO prostate.10
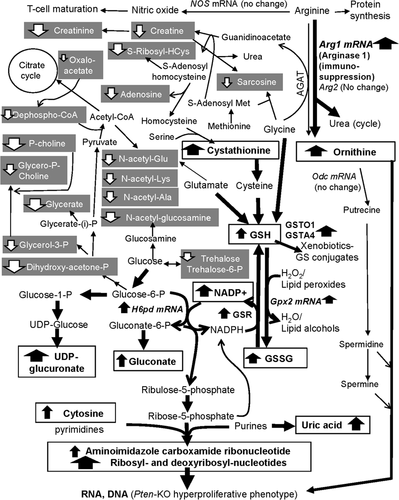
In addition to diversion from creatine/creatinine production, arginine was likely shunted away from nitric oxide synthesis (T cell maturation) to allow arginase to produce the polyamine precursor ornithine (part of the urea cycle). The mRNA level for Arg1 (arginase 1) gene was much increased in our macromolecular omics analyses of Pten-KO prostate,10 consistent with and supportive of infiltration of myeloid-derived (immune) suppressive cells (MDSC) and inflammatory cells.9 Unfortunately, our metabolomics platform failed to detect any polyamines or their further metabolites (no detection in Table S1).
3.3 Comparison with reported metabolomics datasets, and human relevance
As a measure of cross validation, we compared our data set with the metabolomics analysis reported by Zabala-Letona et al. of Pten-KO mouse prostate.16 They reported that the most prominently increased metabolites were higher-order polyamines, such as N-acetyl-spermidine and N-acetyl-spermine, which were not detected by our platform (Table S1). Four metabolites (IDP, ADP, uric acid, and cholesteryl sulfate) matched our panel of 20 increased metabolites (Table 1). In contrast to our finding of elevated abundance of NADP+ and glutathione (GSH), they reported decreased levels of both metabolites in the Pten-KO prostate tumors. Among the 24 decreased metabolites, they reported five that matched the same change trajectory, that is, N-acetylglucosamine, creatinine, phosphorylcholine, adenosine, and sn-glycerol-3-phosphoate (Table 1). However, they reported increased levels of glycerate, hydroxyphenylpyruvate, and N-acetyl-l-alanine in the Pten-KO prostate. These discrepancies could be caused by the difference in the mass spectrometer type (QTRAP 5500 mass spectrometer (ours) vs. Agilent 6520 QToF mass spectrometer16), as well as the prostate tissue sample acquisition and extract preparation methods.
Regarding reported metabolite changes in human PCa specimens, NADP+, ornithine, glutathione, adenosine, and N-acetyl-glutamate were found to change in the same directions as in our Pten-KO mouse data set (Table 1). On the other hand, uric acid, GSSG, N-acetylglucosamine, glycerate, and sn-glycerol-3-phosphate did not change in the same patterns between human PCa and our profiled mouse Pten-KO prostate (Table 1). In particular, both the human PCa datasets from Jung et al.17 and Ren et al.18 could not detect ATP and most of other high-energy phosphorylated nucleotides (e.g., dGTP, dGDP, IDP, ADP, etc.). Previously, many studies failed to show the consistent extraction of high-energy phosphorylated compounds (e.g., ATP and NADPH) known to be susceptible to degradation.19 A mixture of acetonitrile:methanol:water (40:40:20) with 0.1 M formic acid and subsequent neutralization with ammonium bicarbonate was considered as an effective solvent system for both quenching and extraction.19 The solvent systems used for their metabolite extractions (Ren paper: formic acid, ammonium bicarbonate, and methyl tert-butyl ether18; Jung paper: water/ethanol/dichloromethane/acetonitrile17) could be suboptimal for high-energy compounds.
3.4 Aqueous metabolome of TRAMP NECa and comparison with Pten-KO prostate
In the TRAMP NECa, our aqueous metabolomics platform detected increased levels of cystathionine, GSH and GSSG, and purine and pyrimidine nucleotides of both ribose and deoxyribose, yet without a significant change of ornithine and even a decrease of cholesteryl sulfate over their paired counterpart WT prostate (Tables 1 and S3). We detected decreased levels in the TRAMP NECa of adenosine, creatine and creatinine, N-acetyl-glucosamine, glycerate, phosphorylcholine (PC), and glycerophosphocholine (GPC), as in the Pten-KO prostate. These concordant changes in both the Pten-KO and the TRAMP mouse prostate cancer models suggested that these metabolites could play common roles in prostate carcinogenesis, or the changes could be an indication of the disruption of normal prostate metabolic functions. An earlier study using (1)H-NMR showed that in TRAMP mice of 28 weeks of age, reduced tumor levels of citrate (49%), choline (33%), PC (57%), GPC (66%), and glycerophosphoinositol (61%) were observed relative to age-matched WT prostate (p < 0.05).20
However, in contrast to the Pten-KO prostate, prominent increases of many amino acids and their metabolites were detected in the TRAMP NECa (Table S3). Indeed, pathway enrichment analyses (Table S4) showed that five of six significantly (p < 0.05) upregulated metabolic pathways were related to the metabolisms of methionine; glycine and serine; homocysteine degradation; aspartate; and glutamate. The other was purine metabolism.
Figure 3A showed a clear separation of the malignant TRAMP NECa from the WT prostate specimens by supervised PLS-DA of the variance of the differential metabolites between TRAMP (red triangles) and paired WT mouse prostate tissue (green crosses). The heatmap of the top 25 differential metabolites (18 more abundant, seven less abundant than WT) (Figure 3B) showed distinctive clustering of the metabolites between NECa and the normal prostate. Location-based metabolite sets MESA predicted that the altered TRAMP metabolic pathways were mainly associated with peroxisome and lysosome subcellular organelles and the implicated organs were related to NE signaling and innervations (nerve cells; neuron; brain; prostate, pancreas, heart, and muscles) (Figure 3C).
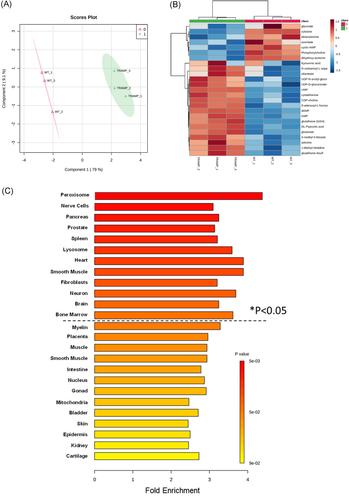
Figure 4 mapped the majority of detected metabolite changes (bold for increased in TRAMP NECa; shaded white for decreased in TRAMP NECa) within the interconnected metabolic pathways, shunts, and cycles. TRAMP NECa appeared to shunt glucose away from glycolysis and citrate cycle, even more so than the Pten-KO prostate. Instead, glucose was channeled toward the glycosylation (increased UDP-glucose) and glucuronidation (increased UDP-glucuronate) pathways. As in the Pten-KO prostate, the TRAMP NECa increased GSH and GSSG, in association with increased cystathionine to increase cysteine, and shunting glycine away from degradation (i.e., decreased creatine and creatinine). Several distinctions from Pten-KO prostate were noteworthy. Foremost, the elevated pools of many amino acids would undoubtedly provide ample substrates for protein synthesis. Besides supporting protein synthesis, the extraordinarily elevated aspartate (Asp) and asparagine (Asn) and elevated carbamoyl-Asp might fuel the de novo syntheses of uracil and subsequent conversion to uridine, cytidine, and the further downstream ribosyl and deoxyribosyl pyrimidine nucleotides for RNA and DNA syntheses. In addition, the increased S-adenosyl-methionine (as well as its de-methylated product S-adenosyl-homocysteine) was interpreted as providing abundant methyl donor for the methylation epigenetic regulations of DNA and other macromolecules. An earlier study had shown DNA hypermethylation of poorly differentiated TRAMP tumors.21 Furthermore, in the TRAMP NECa, elevated levels of UDP-ethanolamine and UDP-choline plus depleted levels of ethanolamine and choline and choline phospho intermediary metabolites might be consistent with potential de novo synthesis of phosphatidyl lipids required for cell membranes in the malignant cells. Moreover, TRAMP NECa decreased arginine degradation (urea level) and did not increase ornithine production, nor uric acid but increased a purine degradation product allantoate which fell further downstream of uric acid.
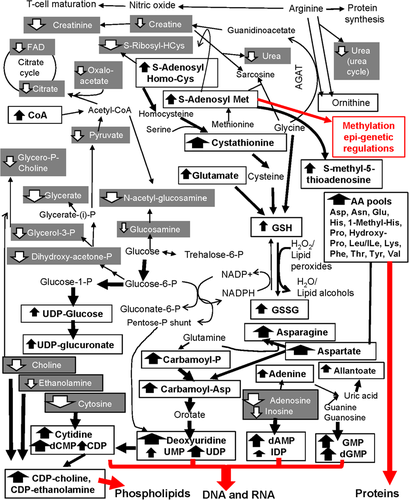
4 DISCUSSION
As PTEN deficiency leads to AKT-mammalian target of rapamycin (mTOR) activation in prostate driving proliferation and transformation of epithelial lesions,22 it was not surprising that increased purine/nucleotides metabolism became the most prominent feature in our aqueous metabolome outcome for Pten-KO prostate (Tables 1 and 2). The metabolisms of purine and pyrimidine are crucial to support the synthesis of nucleic acids.23, 24 Purine synthesis has been reported to be modulated by mTORC1 downstream of the PI3K/PTEN/AKT pathway or vice versa.24, 25 Similarly, Pten loss in mouse embryonic fibroblast (MEF) cells leads to glutamine to cascade through the de novo pyrimidine synthesis.26 The faster-growing transformed cells in the Pten-KO prostate have a high demand for nucleotides for intensive synthesis of DNA and RNA. The transcriptomic profiling indicated greater expression of many transporters, likely enabling the greater fluxes of these needed nucleotides. The second prominent feature was increased glutathione metabolism in the Pten-KO prostate (Table 2; Figure 2). Elevated glutathione and its metabolisms are associated with a proliferative response and tumor progression in various types of tumors.27 Serum cystathionine was higher in prostate cancer patients who developed rapid biochemical recurrence compared to age-matched recurrence-free patients.28 Glutathione is an antioxidant that functions as a free radical scavenger by detoxifying hydrogen peroxide and lipid peroxides through GPXs, and we have reported the specific elevation of Gpx2 mRNA in the Pten-KO prostate in microarray transcriptomics.10 Glutathione is synthesized from glutamate, cysteine, and glycine (Figure 2). In our network analysis, the metabolites in glutathione metabolism were increased (i.e., GSH and GSSG as well as cystathionine, which converted to key amino acid cysteine for GSH synthesis), while those in glycine, serine, and threonine metabolism and alanine, aspartate, and glutamate metabolism were decreased in the PTEN-KO mutant prostate (Table 2; Figure 2), consistent with a tenet that these building blocks of glutathione were shunted toward more synthesis.27 Important for the synthesis of glutathione, the trans-sulfuration pathway is involved in providing sulfur from methionine through cystathionine and eventually to cysteine.29, 30 Consistent with our finding, GSSG in Pten-null mouse liver was found increased by twofold.31 The elevated glutathione metabolism in conjunction with increased expression of enzymes using GSH as a substrate (GSTO1, GSTA4 proteomics, Gpx2 mRNA by microarray)10 might reflect greater redox flux and cytoprotection permissive for a tumor cell growth advantage.18, 31 These two metabolic upregulation changes observed in the Pten-KO prostate were also observed in the TRAMP NECa, consistent with enhanced redox protection for proliferation of the malignant cells which required ample provision of purine and pyrimidine nucleoside/nucleotide substrates for their DNA and RNA syntheses.
Metabolisms of amino acids manifested a major distinction point between Pten-KO and TRAMP carcinogenesis. Arginine is a highly versatile amino acid and is associated with several biological processes beside synthesis of proteins, including nitric oxide (NO) and T cell activation/maturation. As a key arginine catabolism product and polyamine precursor, ornithine was significantly increased in human prostate cancer tissues from radical prostatectomy compared to normal adjacent tissue.17 Ornithine is a key component of the urea cycle and it shuttles between the cytosol and mitochondria. Arginase-1 enzyme in the cytosol converts arginine to urea and ornithine, which can be decarboxylated to generate putrecine and subsequently to higher polyamines. The reshunting of arginine metabolism in the Pten-KO prostate was indicated by elevated abundance of ornithine and a decreased production of creatine and its dehydration product creatinine (Figure 2; Table 2) as a likely result of decreased arginine:glycine aminotransferase flux. Microarray transcriptomic analyses detected increase Arg1 mRNA level, without change of Arg2 or Odc mRNA.10 The increased Arg1 mRNA expression and increased ornithine level in the Pten-KO prostate were consistent with the increased infiltrating MDSC cells in this model for an immune-suppressive TME as detailed by Hong Wu group9 and confirmed by our macromolecular omics.10 In contrast to no change or generally decreased amino acid levels in the Pten-KO prostate, the TRAMP NECa contained elevated pools of many amino acids and lacked indications for greater degradation of arginine (decreased urea, no change in ornithine) (Table S3; Figure 4). In either model, we failed to detect polyamines. However, increased synthesis of polyamines was recently reported as the most prominent feature of metabolomics change in the Pten-KO prostate by Zabala-Letona et al.16 They attributed such increases to mTORC1-driven rewiring of methionine metabolism to increase the polyamine metabolic flux. The discrepancy between our profiling outcome and theirs could be the differences in the metabolome platforms, among many possibilities.
As far as relevance to metabolite changes in human prostate cancer is concerned, increased abundance of hydrophobic cholesteryl fatty acid esters (e.g., oleate and stearate)32 as well as the more hydrophilic cholesteryl sulfate33 have been reported as a distinguishing small-molecule signature of aggressive human prostate cancer. Cholesteryl sulfate was found to increase significantly in the Pten-KO prostate tumors than the WT prostate; but not in the TRAMP model using the same detection platform (Tables 1 and S3), highlighting another crucial metabolite difference between these two models. Unfortunately, we failed to detect any cholesterol itself or its fatty acid esters and all lipid classes. We speculate that the methanolic solvent used to prepare the tissue extracts in our protocol was likely more adept for extracting a polar cholesteryl entity such as the sulfate form than the nonpolar fatty acid esters.
Although in vitro studies have suggested PTEN loss can lead to increased de novo lipogenesis through inducing sterol receptor element binding protein (SREBP) and fatty-acid synthase (FASN),34 the reduced choline intermediates glycerophosphocholine and phosphorylcholine did not support increased biosynthesis of phosphatidylcholine lipids in the Pten-KO prostate (Table 2; Figure 2). Our transcriptomic profiling had reported either no change or decreased mRNA abundance for genes coding for enzymes and proteins involved in fatty acids and lipid synthesis or cholesterol metabolism, such as Hmgcs 1 and 2 (3-hydroxy-3-methyglutaryl-coenzyme A synthase 2), Cav1 (caveolin 1), Fgl1 (fibrinogen like 1), Prom2 (prominin 2), Fads1 (fatty acid desaturase 1), Pnliprp1 (pancreatic lipase related protein 1), Vldlr (very low-density lipoprotein receptor).10 Future work may employ lipidomics to study the hydrophobic sterols, lipids, and fatty acids, which were not detectable by our current platform. In the TRAMP NECa, while the choline and choline intermediary metabolites were decreased as in the Pten-KO prostate, the elevated UDP-choline and UDP-ethanolamine were likely conducive for increased de novo phospholipid syntheses in the aggressively growing NECa (Figure 4).
Many cancers prefer the less efficient but faster aerobic glycolysis (Warburg effect) to generate energy by converting glucose to lactate to support cancer cell growth and proliferation under hypoxic TME, while the differentiated normal cells mainly rely on efficient but slow mitochondrial oxidative phosphorylation to maintain cellular energetics.35 In previous collaborative work, we have shown that an increase in aerobic glycolysis in the Pten-TP53 double deficient prostate cancer cells through a specific elevation of hexokinase-2 (HK-2) isoform, but Pten-KO alone had minimal impact on HK-2 expression or glycolysis.36 In our current metabolomic analysis, we found that the intermediate metabolites of glycolysis and pyruvate metabolisms, including oxaloacetate and dihydroxyacetone phosphate, were modestly decreased in the Pten-KO prostate (Figure 2; Table S1), therefore, not supporting increased glycolytic energy metabolism. Rather the metabolomics outcome implicated an overall reshunting of glucose to pentose phosphate cycle to sustain the generation of NADPH for GSH/GSSG redox, pentose moieties for purine and pyrimidine nucleotides, and to glucuronidation Phase II detoxification activities. The TRAMP NECa shared similar downregulation of glycolysis-citrate cycle metabolism (Figure 4), in spite of the onco-driver differences from the Pten-KO model.
4.1 Limitations
The major caveat for the employed metabolome platform was a bias for hydrophilic metabolites, excluding almost all lipids, fatty acids, and sterols and their hydrophobic metabolites. Another weakness was the lack of anatomical (different prostate lobes) and cell-type distribution information of the metabolites due to the homogenization of each tissue sample. Future efforts should include lipodomics for the hydrophobic metabolites, as well as imaging MS approaches that provide metabolite mapping resolution to cellular or subcellular organelle level on tissue microscopic slice.
5 CONCLUSIONS
In spite of the limitations of the extraction and analytical platform, the aqueous metabolomic change patterns in Pten-KO prostate suggested a significant reshunting of glucose metabolism, GSH synthesis, and redox and arginine to ornithine catabolism for increased proliferation of transformed epithelial cells and immunosuppression. The Pten-KO model exhibited some unique shared metabolism signature with human prostate cancer such as cholesterol sulfate that was decreased in the TRAMP model. While sharing similarities in greater pools of cystathionine and GSH/GSSG redox pair, and of ribosyl and deoxyribosyl nucleotides for nucleic acid syntheses, the TRAMP model stood out in the elevated pools of amino acids for protein synthesis and methylation epigenetic regulations, plus a likelihood of increased de novo synthesis of phospholipids. The metabolomics datasets from these two experimental models of prostate carcinogenesis are accessible to readers for data mining and interpretation.
ACKNOWLEDGMENTS
The authors thank Dr. Christopher J. Kemp and Mr. Kay Gurley at the Fred Hutchinson Cancer Research Center for supplying the frozen bio-specimens; Dr. Su-Ni Tang at Texas Tech University Health Sciences Center for help on tissue processing. This study has been supported, in part, by R01CA172169 grant (to J. L.) and R21CA218774 grant (to J. L.) from US National Cancer Institute and Penn State College of Medicine Start-up fund (to J. L.).
CONFLICT OF INTEREST
The authors declare that there are no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data that supports the findings of this study are available in the supplementary materials of this article.