Genetic polymorphisms at 19q13.33 are associated with [−2]proPSA (p2PSA) levels and provide additional predictive value to prostate health index for prostate cancer
Da Huang, Xiaohao Ruan, and Yishuo Wu contributed equally to this study.
Abstract
Background
Prostate health index (phi), a derivative of [−2]proPSA (p2PSA), has shown better accuracy than prostate-specific antigen (PSA) in prostate cancer (PCa) detection. The present study was to investigate whether previously identified PSA-associated single nucleotide polymorphisms (SNPs) influence p2PSA or phi levels and lead to potential clinical utility.
Methods
We conducted an observational prospective study with 2268 consecutive patients who underwent prostate biopsy in three tertiary medical centers from August 2013 to March 2019. Genotyping data of the 46 candidate genes with a ± 100 kb window were tested for association with p2PSA and phi levels using linear regression. Multivariable logistic regression models were performed and internally validated using repeated tenfold cross-validation. We further calculated personalized phi cutoff values based on the significant genotypes. Discriminative performance was assessed using decision curve analysis and net reclassification improvement (NRI) index.
Results
We detected 11 significant variants at 19q13.33 which were p2PSA-associated independent of PCa. The most significant SNP, rs198978 in KLK2 (Pcombined = 5.73 × 10−9), was also associated with phi values (Pcombined = 3.20 × 10−6). Compared to the two commonly used phi cutoffs of 27.0 and 36.0, the personalized phi cutoffs had a significant NRI for PCa ranged from 5.23% to 9.70% among men carrying variant types (all p < .01).
Conclusion
Rs198978, is independently associated with p2PSA values, and can improve the diagnostic ability of phi for PCa using personalized cutoff values.
1 INTRODUCTION
Prostate-specific antigen (PSA) is the most widely used biomarker for screening of prostate cancer (PCa). However, its poor ability to distinguish aggressive PCa from indolent diseases leads to a massive overdiagnosis.1 To solve these clinical predicaments, prostate health index (phi) derived from total PSA (tPSA), free PSA (fPSA), and [−2]proPSA (p2PSA), has been introduced. It has shown better accuracy in predicting PCa and clinically significant PCa (csPCa) than PSA, especially in patients with tPSA between 2.0 and 10 ng/ml.2-4 Our previous study also reported that phi outperformed PSA among Chinese men with tPSA > 10 ng/ml.4
Mature PSA is activated from precursor PSA (proPSA) after cleavage by human kallikrein-2 (hK2, also known as kallikrein-related peptidase 2 [KLK2]).5 Alternatively, p2PSA, another inactive truncated form of proPSA, once formed, is not cleaved by hK2.6 Sharing the same pathway, the increased synthesis of p2PSA may be related to decreased cleavage by hK2 in PCa cells.7
In the recent decade, plenty of single nucleotide polymorphisms (SNPs) have been reported to be associated with tPSA or fPSA levels in genome-wide association studies (GWAS) among PCa cases and controls.8-18 These variants have potentially important clinical implications if they are independently associated with biomarker levels (but not with PCa risk). For example, the genetic-adjusted PSA based on these SNPs can improve predictive performance by applying personalized cutoff values.17-20 Furthermore, previous non-GWAS association studies reported several genetic variants associated with serum hK2 levels.21-24 Thus, the correlative genetic variants may also affect the levels of serum p2PSA. However, the genetic or inherited influence on p2PSA levels is poorly understood and has never been reported.
The objective of this study is to investigate whether the established tPSA- and fPSA-associated SNPs are also independently associated with p2PSA or phi levels. To implement the results to clinical translational practice, we also assessed the clinical utility of personalized phi cutoffs adjusted by the genetic variants.
2 MATERIALS AND METHODS
2.1 Study population
In this prospective, observational study, we established a multicenter prostate biopsy cohort from three tertiary medical centers (Shanghai Ruijin Hospital, Shanghai Huashan Hospital, and Shanghai Cancer Center) in Shanghai, China. A total of 2268 patients who underwent initial prostate biopsies were consecutively enrolled from August 2013 to March 2019. This is a subset of the entire cohort with the information of genotyping array data.
The indications for prostate biopsy were the same across different centers: (1) a tPSA level > 10.0 ng/ml; (2) a tPSA level > 4.0 ng/ml with confirmation after 2–3 months; (3) %fPSA < 0.16 when patients had a tPSA level > 4.0 ng/ml; and (4) the presence of suspicious lesions detected by digital rectal examination (DRE), ultrasound or magnetic resonance imaging (MRI) at any level of tPSA. The phi calculation was not used in clinical decision-making. MRI was not used for biopsy decision-making in patients with a tPSA level > 4.0 ng/ml.
Blood samples were collected for genotyping and the measurement of tPSA, fPSA, and p2PSA before biopsies on the same day in a central certified lab per the study protocol. Transrectal ultrasound-guided biopsies were performed using a 10- to 14-core scheme. All biopsy specimens were independently examined and graded by two experienced pathologists in the department of pathology at each hospital. Based on the new Gleason grading system in 2015, csPCa was defined as PCa with Gleason Grade ≥ 3 (as known as Gleason Scores 4 + 3, 8, 9, and 10).25
Patients were excluded from the present study if serum antigen levels (tPSA, fPSA, or p2PSA) were unable to be tested due to poor serum sample quality. The study was approved by the Institutional Review Board of each hospital, and written informed consent was obtained from each participant.
2.2 SNP selection and genotyping
Genotyping was performed using the Illumina Asian Screening Array (ASA) BeadChip platform covering ~660k variants across the genome. Using the combined data of the 1000 Genomes project and HapMap3 data as reference, genotypes of SNPs that were not genotyped were imputed by IMPUTE version 2 software.26 A posterior probability of >0.90 was applied to call genotypes during imputation and the same quality control procedure for excluding genotyped SNPs was applied to imputed SNPs.
We selected 81 tPSA- or %fPSA-associated SNPs in 46 candidate genes and their intergenic regions from previous GWAS studies (Table S1).8-18 Genotyping data of the 46 candidate genes with a ± 100 kb window was extracted. SNPs were excluded if they had: (1) genotype call rate < 90% (n = 6833); (2) minor allele frequency (MAF) < 0.01 (n = 5989); or (3) p < .05 for the Hardy–Weinberg Equilibrium (HWE) test (n = 1250).
2.3 Quantitative association analysis
The derivative variable phi was calculated as follows: (p2PSA/fPSA) × √tPSA. All the serum indices (including tPSA, fPSA, p2PSA, and phi) were logarithm- (log-) transformed before quantitative association analyses. We randomly divided the entire cohort into two groups and performed a two-stage association study by using PLINK software (version 1.07).27 At each stage, multivariable linear regression analyses were used to evaluate the effect (beta values [β] and standard error) of each SNP on p2PSA or phi levels after adjusting for age and biopsy outcomes (PCa/non-PCa), and an additive model for allelic effect was assumed. The meta-analysis of the two subgroups was performed and P-combined values were estimated using the random-effects model. A p < .05 was considered statistically significant after the Bonferroni correction. Using the CHB population (Han Chinese in Beijing, China) as reference, the p value results and linkage disequilibrium patterns for SNPs in a region of interest were measured and visualized by LDassoc and LDmatrix Tool.28, 29
2.4 Genetic-adjusted p2PSA/phi values and personalized cutoff values
The genetic correction of p2PSA levels was calculated as follows. First, subtract the log-transformed original p2PSA levels with the estimated relative genetic effects of the p2PSA-associated SNPs (β values in multivariable linear regression models); Second, take reverse log-transformation as the final genetic-adjusted p2PSA values. The genetic-adjusted phi values were calculated in the same way.
Using the three commonly used phi cutoffs (27, 36, and 55)30, 31 as reference, personalized cutoff values for phi were calculated for each subject. The personalized cutoff values were determined by keeping the same level of specificity in patients with or without genetic variants.
2.5 Statistical analysis
For baseline characteristics, continuous variables were compared using Student's t-test after log-transformation or Cuzick's test for trend across ordered groups (SNP genotypes: wild type, heterozygous, and homozygous variant type). Categorical variables were compared using Fisher's exact test. Ordinal variables were compared using Cochran–Armitage tests for trend. To evaluate whether an SNP of interest provides additional predictive value for PCa and csPCa, we conducted multivariable logistic regression (LR) models including age, phi values, and SNP genotypes as covariates, and calculated adjusted odds ratios (aOR), 95% confidence intervals (95% CI) and p values for each covariate. The predictive abilities of phi, genetic-adjusted phi, and LR models were evaluated using the area under the receiver operating characteristic curve (AUC) and decision curve analysis (DCA).32 All the LR models were corrected for overfit by using repeated tenfold cross-validation (200 replications) before comparison. The net reduction in the number of biopsies between different models was also evaluated by DCA. Comparing to a commonly used phi cutoff, the performance of the personalized cutoff was evaluated using the net reclassification improvement (NRI) index.33 A Z-test was used to test for the null hypothesis of NRI = 0.
All statistical analyses were performed using Stata 15.1 Special Edition (StataCorp). A two-tailed p < .05 was considered statistically significant.
3 RESULTS
The baseline characteristics and biopsy outcomes in the entire cohort and subgroups of two stages are shown in Table S2. Based on our exclusion criteria, 14 patients were excluded due to the failure of serum antigen test, and 2254 patients with sufficient phenotypic and genotypic information were included for further association analyses. A total of 974 (43.2%) patients had PCa documented on biopsy, and 529 (23.5%) had csPCa. There were no significant differences in the baseline and outcomes between subjects in the two stages (All p > .05).
After quality control of genotyping results and imputed SNPs, a total of 15,433 SNPs in 2254 subjects was used for further association analyses. We evaluated the quantitative association between each individual SNP on p2PSA and phi levels at each stage adjusted for age and biopsy outcomes (Table 1). When the two stages were combined, we found that 11 SNPs in chromosome 19 were significantly associated with p2PSA levels at a Pcombined value < 3.24 × 10−6 (Bonferroni corrected significant level = 0.05/15,433). Among the 11 significant variants, there were 8 SNPs in the KLK2 gene and 3 in the KLKP1 gene. All of the 11 SNPs were in the same linkage disequilibrium (LD) region, with r2 values ranging from 0.907 to 0.969 (Figure 1). The most significant SNP, rs198978 (G > T) in the KLK2 gene at 19q13.33, was also the only SNP that was significantly associated with phi values (Pcombined = 5.73 × 10−9 and 3.20 × 10−6 for p2PSA and phi, respectively, Table 1). These indicated that the SNPs were independently associated with p2PSA and phi levels regardless of the disease status (PCa/non-PCa).
| SNP | Region | Position | Alleles | Gene | Consequence | Stage | log-transformed p2PSA | log-transformed phi | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | pa | Pcombined | β (SE) | p | Pcombined | |||||||
| rs6070 | 19q13.33 | 51,380,110 | A/T | KLK2 | Intron variant | 1 | 0.13 (0.03) | 3.09E−05 | 1.70E−08 | 0.08 (0.02) | 4.73E−04 | 1.20E−05 |
| 2 | 0.12 (0.03) | 1.63E−04 | 0.06 (0.02) | .008 | ||||||||
| rs8105275 | 19q13.33 | 51,380,445 | T/G | KLK2 | Intron variant | 1 | 0.14 (0.03) | 2.01E-05 | 4.35E−08 | 0.08 (0.02) | 5.57E−04 | 2.50E−05 |
| 2 | 0.11 (0.03) | 5.71E−04 | 0.06 (0.02) | .013 | ||||||||
| rs198974 | 19q13.33 | 51,380,602 | G/T | KLK2 | Intron variant | 1 | 0.13 (0.03) | 2.45E−05 | 1.67E−08 | 0.08 (0.02) | 2.13E−04 | 1.06E−05 |
| 2 | 0.12 (0.03) | 1.96E−04 | 0.06 (0.02) | .013 | ||||||||
| rs198975 | 19q13.33 | 51,380,815 | T/G | KLK2 | Intron variant | 1 | 0.13 (0.03) | 2.30E−05 | 2.71E−08 | 0.08 (0.02) | 1.91E−04 | 1.13E−05 |
| 2 | 0.11 (0.03) | 3.23E−04 | 0.06 (0.02) | .015 | ||||||||
| rs8108845 | 19q13.33 | 51,381,083 | C/G | KLK2 | Intron variant | 1 | 0.14 (0.03) | 1.16E−05 | 3.25E−08 | 0.08 (0.02) | 2.09E−04 | 1.49E−05 |
| 2 | 0.11 (0.03) | 6.71E−04 | 0.06 (0.02) | .017 | ||||||||
| rs198976 | 19q13.33 | 51,381,126 | T/C | KLK2 | Intron variant | 1 | 0.13 (0.03) | 2.22E−05 | 2.54E−08 | 0.08 (0.02) | 1.79E−04 | 1.21E−05 |
| 2 | 0.11 (0.03) | 3.15E−04 | 0.05 (0.02) | .016 | ||||||||
| rs198977 | 19q13.33 | 51,381,777 | T/C | KLK2 | NC transcript/Missense/3ʹ-UTR variant | 1 | 0.13 (0.03) | 1.47E−05 | 1.23E−08 | 0.08 (0.02) | 2.84E−04 | 1.13E−05 |
| 2 | 0.11 (0.03) | 2.23E−04 | 0.06 (0.02) | .011 | ||||||||
| rs198978 | 19q13.33 | 51,383,072 | T/G | KLK2 | NC transcript/3ʹ-UTR variant | 1 | 0.13 (0.03) | 3.40E−05 | 5.73E−09 | 0.08 (0.02) | 1.76E−04 | 3.20E−06 |
| 2 | 0.12 (0.03) | 4.98E−05 | 0.06 (0.02) | .005 | ||||||||
| rs16987929 | 19q13.33 | 51,385,253 | G/A | KLKP1 | Downstream variant | 1 | 0.13 (0.03) | 1.93E−05 | 1.22E−08 | 0.08 (0.02) | 4.84E−04 | 1.49E−05 |
| 2 | 0.11 (0.03) | 1.72E−04 | 0.06 (0.02) | .009 | ||||||||
| rs8103659 | 19q13.33 | 51,385,402 | G/A | KLKP1 | NC transcript variant | 1 | 0.13 (0.03) | 4.58E−05 | 3.40E−08 | 0.07 (0.02) | 6.82E−04 | 1.77E−05 |
| 2 | 0.11 (0.03) | 2.14E−04 | 0.06 (0.02) | .008 | ||||||||
| rs198958 | 19q13.33 | 51,388,878 | A/T | KLKP1 | Intron variant | 1 | 0.10 (0.03) | 1.78E−03 | 2.74E−06 | 0.06 (0.02) | 7.20E−03 | 2.84E−04 |
| 2 | 0.11 (0.03) | 4.96E−04 | 0.06 (0.02) | .015 | ||||||||
- Abbreviations: 3ʹ-UTR, three prime untranslated region; β, regression coefficient; p2PSA, [−2]proPSA; log, logarithm; NC, noncoding; phi, prostate health index; SE, standard error.
- a P values were estimated using multivariate linear regression on logarithm-transformed p2PSA or phi values after adjusting for age and biopsy outcomes (prostate cancer or not), and an additive model for allelic effect was assumed. Pcombined values were estimated using random-effects meta-analysis across two-stage association studies.
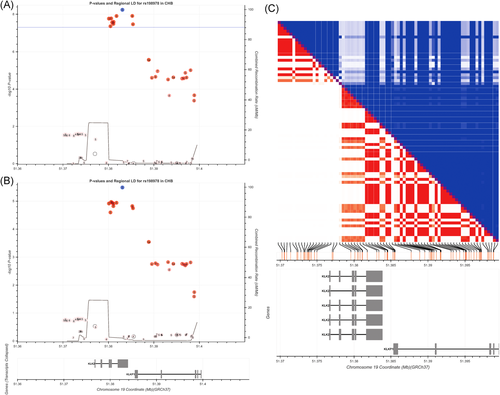
We then performed a stratified analysis based on rs198978 genotypes in the entire cohort and patients with tPSA levels of 2–10 ng/ml. In the entire cohort, men carrying heterozygous and homozygous variant types (TT and TG) had significantly higher fPSA, p2PSA, and phi values than those with wild type (all p < .05, Table S3). In addition, p2PSA and phi values both had increased significantly across ordered genotype groups (all p for trend < .001, Table 2). The genotype's effect on p2PSA and phi values remained significant in PCa and non-PCa groups (Table S4). However, no significant difference was observed in age and tPSA values among different rs198978 genotypes (all p > .05, Table S3). Similar results were also found among patients with tPSA levels of 2–10 ng/ml.
| Rs198978 | N | Meana | Meana | PCa, n (%) | PCa vs. non-PCa | csPCa, n (%) | csPCa vs. others | ||
|---|---|---|---|---|---|---|---|---|---|
| p2PSA | phi | aORb (95% CI) | p | aORb (95% CI) | p | ||||
| Entire cohort | |||||||||
| GG | 1478 | 30.3 | 59.3 | 637 (43.1) | 1.00 (Ref.) | 335 (22.7) | 1.00 (Ref.) | ||
| TG | 620 | 42.7 | 71.3 | 275 (44.4) | 0.79 (0.61–1.01) | .064 | 160 (25.8) | 0.98 (0.73–1.33) | .904 |
| TT | 83 | 52.1 | 84.6 | 36 (43.4) | 0.49 (0.27–0.89) | .019 | 21 (25.3) | 0.99 (0.52–1.91) | .985 |
| P for trendc | / | 8.22 × 10−13 | 2.63 × 10−6 | 0.685 | / | / | 0.093 | / | / |
| tPSA 2–10 ng/ml | |||||||||
| GG | 513 | 12.5 | 33.2 | 122 (23.8) | 1.00 (Ref.) | 35 (7.2) | 1.00 (Ref.) | ||
| TG | 217 | 16.0 | 36.5 | 48 (22.1) | 0.71 (0.46–1.10) | .122 | 14 (6.7) | 0.76 (0.38–1.50) | .426 |
| TT | 25 | 20.0 | 40.9 | 5 (20.0) | 0.47 (0.15–1.47) | .192 | 3 (12.5) | 1.39 (0.35–5.46) | .641 |
| P for trendc | / | 3.28 × 10−11 | 9.77 × 10−4 | 0.537 | / | / | 0.705 | / | / |
- Abbreviations: 95% CI, 95% confidence interval; aOR, adjusted odds ratio; csPCa, clinically significant prostate cancer; p2PSA, [−2]proPSA; PCa, prostate cancer; phi, prostate health index; Ref, reference; tPSA, total prostate-specific antigen.
- a Both of the p2PSA levels and phi were log-transformed before calculation and the values presented were back-transformed.
- b Adjusted odds ratios were determined by multivariate logistic regression analysis after adjusting for age and log-transformed phi values.
- c The p values for trend were determined by Cuzick's test.
The association between the loci at 19q13.33 and PCa risk was shown in Table 2. In univariate analysis, no significant difference was observed in the incidence of PCa and csPCa between men with different rs198978 genotypes (GG vs. TG vs. TT, p for trend = .685 and .093 for PCa and csPCa, respectively). Similar results were also observed in multivariable analysis on csPCa risk (all p > .05). However, men carrying variant types showed a mildly lower positive biopsy rate (aORTG = 0.79, 95% CI = 0.61–1.01, p = .064; aORTT = 0.49, 95% CI = 0.27–0.89, p = .019), compared with men carrying homozygous wild type after adjusting for age and phi values.
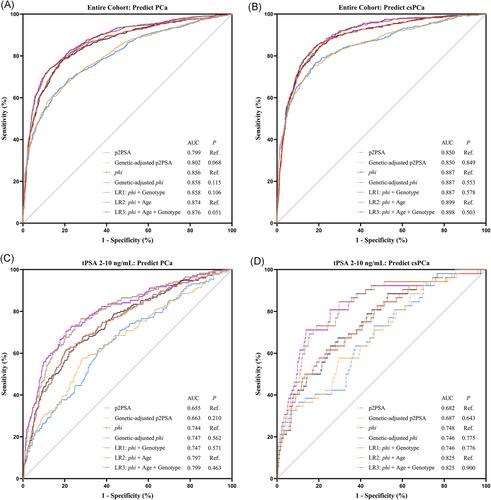
The predictive abilities of different diagnostic models were compared in Figure 2. Briefly, the AUCs of genetic-adjusted p2PSA/phi did not outperform original p2PSA/phi for predicting PCa or csPCa (all p > .05, Figures 2A and 2B). Similar results were found among patients with tPSA levels of 2–10 ng/ml (Figures 2C and 2D). All the LR models did not outperform the base models (LR1 vs. phi, LR3 vs. LR2), which indicated that incorporating the genotypes might not improve the overall discriminating abilities of the original LR models.
After correction for overfit by cross-validation, net benefits of different diagnostic models are shown in Figure 3. In the entire cohort, the LR1 model (phi + rs198978 genotype) could achieve a mildly higher net benefit than the original phi for PCa risk thresholds between 20% and 35% (Figure 3A). For csPCa, the LR1 model had a higher net benefit than the original phi across the entire spectrum of threshold probability (Figure 3B). However, no significant increase in net benefit was observed between the full model (LR3) and LR2 in either the entire cohort or patients with tPSA 2–10 ng/ml (Figures 3C and 3D).
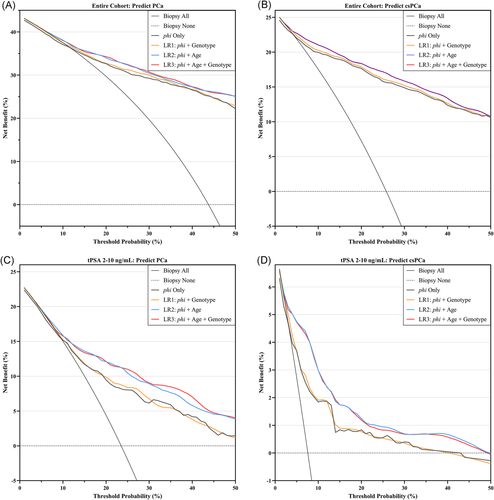
To determine whether rs198978 genotype would provide additional predictive value to phi, we further calculated the personalized phi cutoff values on the basis of three commonly used cutoffs (27, 36, and 55). Among men carrying variant types, using a phi cutoff of 30.9 would cause 5.23% (p = .001) of the total population to reclassify into the correct biopsy decisions, compared with a commonly used cutoff of 27 (Table 3). Similarly, a phi cutoff of 42.2 would perform significantly better than the commonly used cutoff of 36 (NRI = 9.70%, p < .001). Similar results were found when predicting csPCa (all p < .001). However, among patients with tPSA levels of 2–10 ng/ml, no significant difference in discriminability between the personalized and the commonly used cutoffs (all p > .05). By applying the personalized cutoffs to men carrying variant types, higher net reduction in number of biopsies could be achieved at risk thresholds above 15% for PCa (Figures S1A and S1C) and approximately 10% for csPCa (Figures S1B and S1D). However, the personalized cutoffs could not improve the accuracy of the commonly used cutoff of 55, with negative NRI and net reduction in number of biopsies (Table 3 and Figure S1).
| Traditional cutoffs | Wild type (rs198978: GG) | Traditional and Adjusted cutoffsa | Variant types (rs198978: G > T) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | NRI (%) | PNRIb | ||
| Entire cohort: predict PCa | |||||||||||
| 27.0 | 94.5 | 35.4 | 52.7 | 89.4 | 27.0 (Ref.) | 97.7 | 25.8 | 51.2 | 93.5 | 5.23 | 0.001 |
| 30.9 | 95.8 | 35.5 | 54.2 | 91.4 | |||||||
| 36.0 | 88.8 | 58.8 | 62.2 | 87.3 | 36.0 (Ref.) | 93.6 | 46.3 | 58.1 | 90.0 | 9.70 | <0.001 |
| 42.2 | 91.0 | 58.6 | 63.6 | 89.1 | |||||||
| 55.0 | 74.4 | 82.3 | 76.2 | 80.8 | 55.0 (Ref.) | 83.9 | 79.8 | 76.8 | 86.2 | −0.66 | 0.613 |
| 60.1 | 80.7 | 82.4 | 78.4 | 84.3 | |||||||
| Entire cohort: predict csPCa | |||||||||||
| 27.0 | 97.6 | 30.9 | 31.4 | 97.6 | 27.0 (Ref.) | 98.9 | 22.6 | 33.2 | 98.1 | 7.83 | <0.001 |
| 30.6 | 98.3 | 31.0 | 35.7 | 98.0 | |||||||
| 36.0 | 93.4 | 51.4 | 38.4 | 96.0 | 36.0 (Ref.) | 96.7 | 41.5 | 39.1 | 97.0 | 8.02 | <0.001 |
| 41.5 | 95.0 | 51.2 | 43.1 | 96.4 | |||||||
| 55.0 | 86.9 | 75.3 | 53.3 | 94.7 | 55.0 (Ref.) | 90.6 | 73.1 | 56.7 | 95.2 | −0.61 | 0.665 |
| 59.7 | 87.8 | 75.3 | 58.0 | 94.1 | |||||||
| tPSA 2–10 ng/ml: predict PCa | |||||||||||
| 27.0 | 85.2 | 43.5 | 32.0 | 90.4 | 27.0 (Ref.) | 88.7 | 28.0 | 25.7 | 89.8 | 7.80 | 0.099 |
| 30.6 | 81.1 | 43.4 | 28.7 | 89.1 | |||||||
| 36.0 | 68.9 | 69.1 | 41.0 | 87.7 | 36.0 (Ref.) | 71.7 | 55.6 | 31.1 | 87.5 | 8.10 | 0.056 |
| 41.6 | 66.0 | 69.3 | 37.6 | 87.9 | |||||||
| 55.0 | 38.5 | 91.6 | 58.8 | 82.7 | 55.0 (Ref.) | 41.5 | 90.5 | 55.0 | 84.7 | −4.60 | 0.170 |
| 58.1 | 35.8 | 91.5 | 54.3 | 83.6 | |||||||
| tPSA 2–10 ng/ml: predict csPCa | |||||||||||
| 27.0 | 91.4 | 40.7 | 10.7 | 98.4 | 27.0 (Ref.) | 88.2 | 26.5 | 8.7 | 96.6 | 8.07 | 0.208 |
| 30.5 | 82.4 | 40.5 | 9.9 | 96.7 | |||||||
| 36.0 | 71.4 | 65.1 | 13.7 | 96.7 | 36.0 (Ref.) | 70.6 | 53.5 | 10.7 | 95.8 | −0.14 | 0.988 |
| 41.3 | 58.8 | 65.1 | 11.8 | 95.2 | |||||||
| 55.0 | 42.9 | 88.4 | 22.4 | 95.2 | 55.0 (Ref.) | 41.2 | 88.4 | 21.9 | 95.0 | / | / |
| 55.0 | 41.2 | 88.4 | 21.9 | 95.0 | |||||||
- Abbreviations: csPCa, clinically significant prostate cancer; PPV, positive predictive value; NPV, negative predictive value; NRI, net reclassification improvement index; PCa, prostate cancer; phi, prostate health index; Ref, reference; tPSA, total prostate-specific antigen.
- a The adjusted cutoff values were determined by keeping the same level of specificity in patients with or without genetic variants (rs198978: G > T).
- b P values for NRI (PNRI) were estimated by a two-side Z-test for the null hypothesis of NRI = 0.
4 DISCUSSION
In this two-stage tagged association study based on a multicenter cohort, we identified one independent locus at the KLK2 gene in 19q13.33 that was significantly associated with both p2PSA and phi levels but not associated with PCa risk. Men carrying variant types (rs198978: G > T) had significantly higher p2PSA and phi values than those with wild type. No significant association was observed between the detection rate of PCa/csPCa and rs198978 genotype. The adjusted cutoffs of phi outperformed the two commonly used one-size-fits-all cutoffs (27 and 36) in men with variant types. These findings may improve the clinical utility of phi in the detection of PCa and csPCa and help to avoid a large number of unnecessary biopsies.
An elevated biomarker, caused by genetic effects rather than oncogenesis, may decrease diagnostic accuracy due to more false-positive cases. Several previous studies reported the feasibility of incorporating genetic markers into PSA screening.34-37 One of the most commonly used methods was adding significant SNPs and PSA values into a multivariable model. However, all the previous studies demonstrated that adding SNPs to the predictive model would not significantly improve the discriminating abilities of PSA (with only marginal area under receiver operating characteristic curve [AUC] improvement around ~0.02).34-37 Similarly, in the present study, multivariable LR model had a mildly higher net benefit than phi only in the entire cohort (Figure 2). The other method was establishing personalized cutoff values of PSA based on the cumulative effects of significant SNPs.17-20 For example, Gudmundsson et al.18 have identified four PSA-associated markers in a GWAS and calculated a personalized PSA cutoff value based on the distribution of genotypic effect, which showed that ~6%–7% had at least one PSA value reclassified into higher or lower PCa risk categories. We identified only one significant p2PSA-associated locus in the present study. Thus, we did not estimate the cumulative effects, and calculated the adjusted cutoff values by keeping the same specificity of phi across rs198978 genotypes. Compared to the two traditional phi cutoff values of 27 and 36, the net reclassification percentage of our personalized phi cutoffs ranged from 5.23% to 9.70%, which were similar to the results by Gudmundsson et al.18 In summary, when evaluating the genetic adjusted biomarkers, the point estimation method such as personalized cutoff and NRI might be more suitable than the overall estimation including AUC and multivariable regression models.
The underlying molecular mechanism of our findings is still unknown. In the present study, all the identified p2PSA-associated SNPs were at a strong LD block in a region incorporating KLK2 and KLKP1 gene, which both belonged to the kallikrein gene family on chromosome 19. The KLK2 gene encodes hK2 protein, which is the major activating enzyme for proPSA cleavage in vivo.38 The elevation of these truncated proPSA isoforms (such as p2PSA, also known as [−2]proPSA) may be associated with the downregulation of KLK2 gene. Previous studies have revealed that KLKP1 was a pseudogene of kallikreins, of which the expression level was lower in PCa cells than that in normal prostate cells and was regulated by androgen.39, 40 In BPH samples, KLKP1 mRNA levels strongly correlated with PSA mRNA levels.39 However, further studies would be necessary to demonstrate the complex functional mechanism of KLK2 and KLKP1, which might regulate p2PSA expression through androgen receptor.
There were several limitations of this study. First, this is a tagged association study based on 46 candidate genes with a ± 100 kb window. Loci outside these regions might exist. However, this will not change the implementation of our current findings to clinical practice in terms of personalized cutoffs. Second, the sample size of patients with tPSA values of 2–10 ng/ml was relatively small, which might be due to the high likelihood of high-grade PCa in China compared to Western populations.41 However, phi performed better than PSA in patients with higher tPSA levels (>10 ng/ml) in the Chinese population. The application of p2PSA testing is not limited to PSA grey zone in China at this stage. Therefore, we also performed the current analyses in the entire cohort. Third, both the association study and modeling were performed in the same cohort, which might lead to an over-estimate. Although the internal tenfold cross-validation has been performed in the present study, further external validation is needed if applicable. Fourth, the present study may have selection bias because all the three hospitals were located in Shanghai, a large city in East China. However, nearly half of the patients seeking services in these tertiary hospitals were from provinces other than Shanghai.
5 CONCLUSIONS
A locus at 19q13.33 (rs198978) is identified to be associated with p2PSA and phi levels but not PCa risk. Personalized phi cutoff values, based on the genotype, would improve the predictive accuracy of phi in the decision-making for prostate biopsy.
ACKNOWLEDGMENTS
The authors thank all the subjects included in this study. This study was in part supported by grants from the National Natural Science Foundation of China (Grant nos. 81772741, 81972645, and 81972405), Shanghai Rising-Star Program (Grant no. 18QA1402800), the “Chen Guang” project from Shanghai Municipal Education Commission, and Shanghai Education Development Foundation (Grant no. 17CG09), Shanghai Jiao Tong University School of Medicine Gaofeng-Clinical Medicine Grant Support (Grant no. 20181701), and Shanghai Municipal Human Resources and Social Security Bureau (Grant no. 2018052).
CONFLICT OF INTEREST
In the present study, we declare that Beckman Coulter, Inc. provided the tests for tPSA, fPSA, and p2PSA. All the sample collection, data analyses, and manuscript writing were performed by the researchers, independent from Beckman Coulter, Inc. There are no other potential competing interests to be declared.
ETHICS STATEMENT
The study was approved by the Institutional Review Board of Shanghai Ruijin Hospital, Shanghai Huashan Hospital, and Shanghai Cancer Center in Shanghai, China, and written informed consent was obtained from each participant (Central IRB no. KY2016-343, November 24, 2016, version 03).
Open Research
DATA AVAILABILITY STATEMENT
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.




